Marker for pre-judging severe new coronal pneumonia (COVID-19) and product and application thereof
A marker, critically ill technology, applied in recombinant DNA technology, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problems of many detection factors, complex prediction process, and unsatisfactory single prediction effect. To achieve the effect of reducing the fatality rate and simplifying the detection process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1S100A12 as a severe infection marker
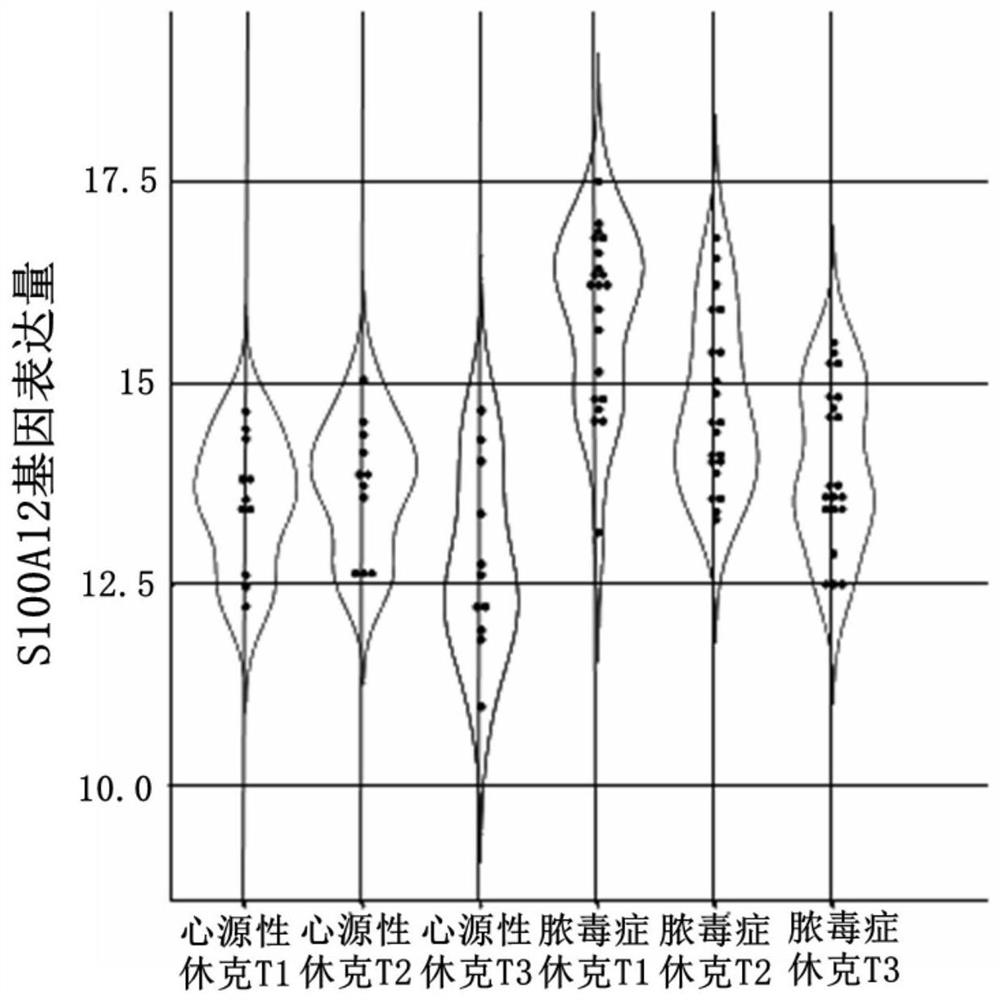
[0050]Collect 13 sets of relevant peripheral blood transcriptome data from the public platform, and conduct in-depth analysis on the gene expression data of 1648 samples. These data include data related to sepsis, data related to influenza, and data related to new coronary pneumonia.
[0051] (1) S100A12 as a marker of severe infection
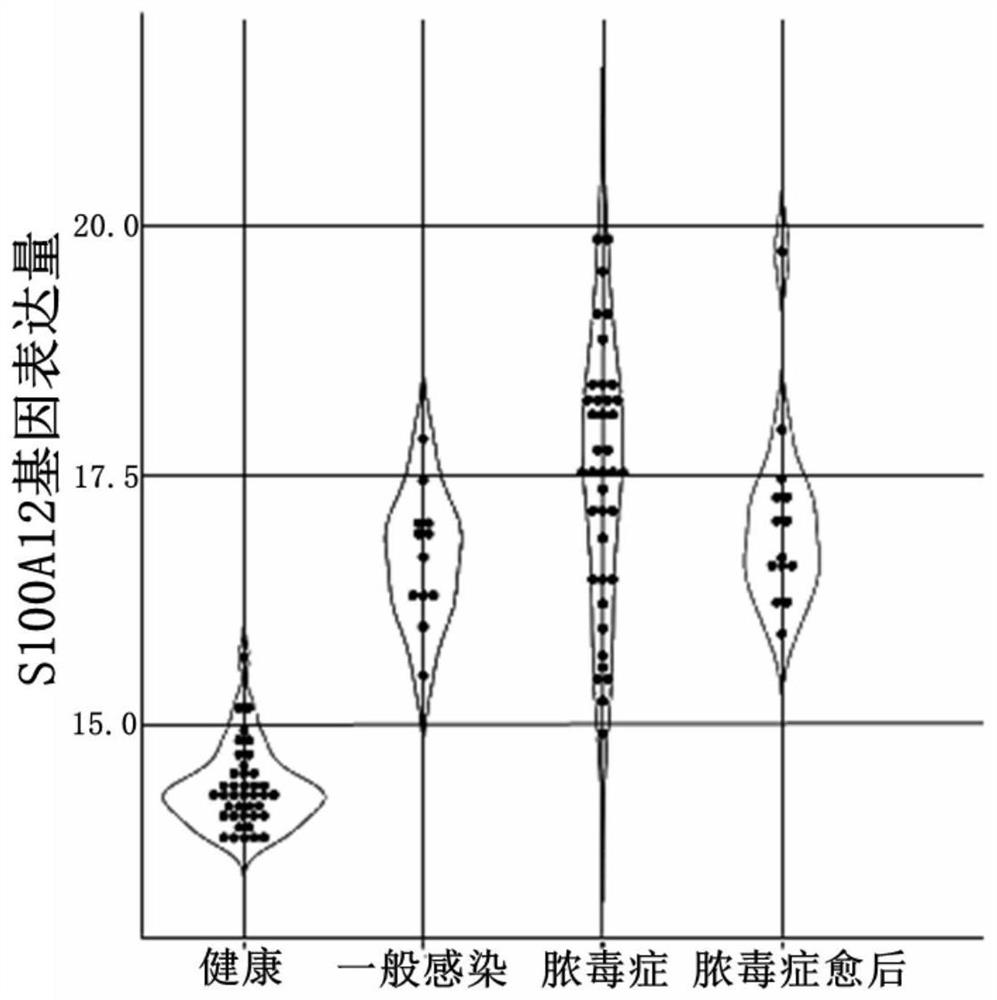
[0052] From figure 1 It can be seen that the expression level of S100A12 (SEQ ID NO.1) is significantly increased in general infection (compared with the healthy group) (p=1.95e-8). In the sepsis group, the expression level of S100A12 further increased (p=0.0074). Therefore, S100A12 can be used as a marker of severe infection.
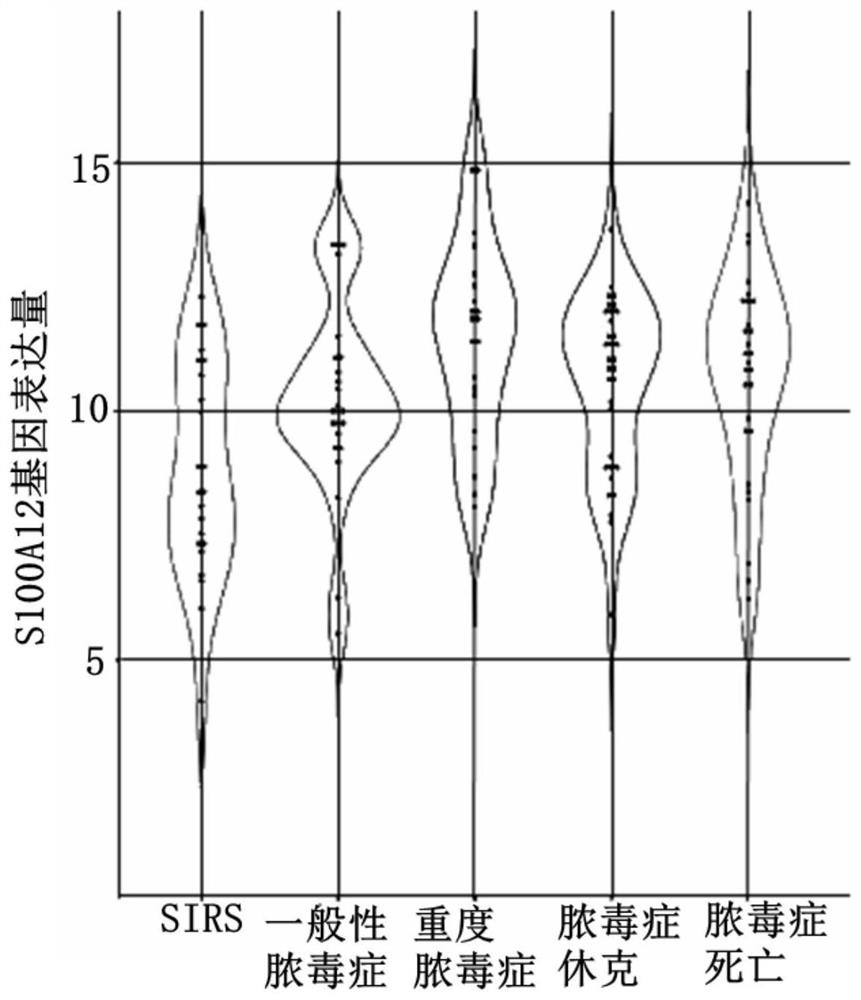
[0053] (2) Correlation between the expression level of S100A12 and the severity of sepsis
[0054] From figure 2 It can be seen that the expression level of S100A12 in the mild sepsis group was significantly higher than that in the SIRS group (ie patients w...
Embodiment 2
[0076] Example 2 A method of predicting severe new coronary pneumonia
[0077] (1) The primer synthesis of the target gene S100A12 and the reference gene B2M was entrusted to Shanghai Sangong. The specific sequence is as follows:
[0078] S100A12-F SEQ ID NO.2 S100A12-R SEQ ID NO.3 B2M-F SEQ ID NO.4 B2M-R SEQ ID NO.5
[0079] (2) Collect 100 μL of venous blood from the subject (50 μL or fingertip blood can also be collected here).
[0080] (3) Mix whole blood with three volumes of lysate (Bitec whole blood lysate). The RNA was purified with a kit (Bitek Whole Blood RNA Purification Kit) and the RNA concentration and quality were detected.
[0081] (4) Reverse transcription kit of Takara, product number RR037A was used for reverse transcription; kit of Thermo Fisher Corporation was used for PCR amplification, product number was A44647100. In this way, the Ct values of the target gene and the internal reference gene were obtained.
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com