Q-PCR detection method for colorectal cancer marker DCD
A detection method, a colorectal cancer technology, is applied in the field of Q-PCR detection of the colorectal cancer marker DCD, which can solve problems such as the unsystematic establishment of the detection method, and achieve the effects of preventing secondary pollution, strong specificity, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1~2
[0037] Reagents used in Examples 1-2
[0038]
[0039]
Embodiment 1
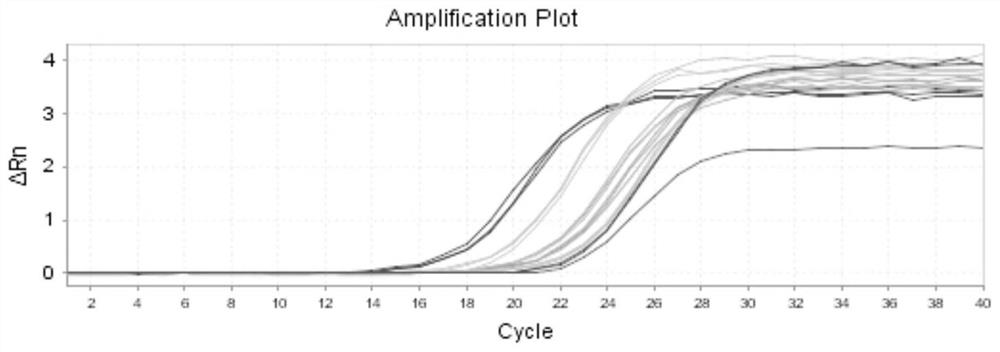
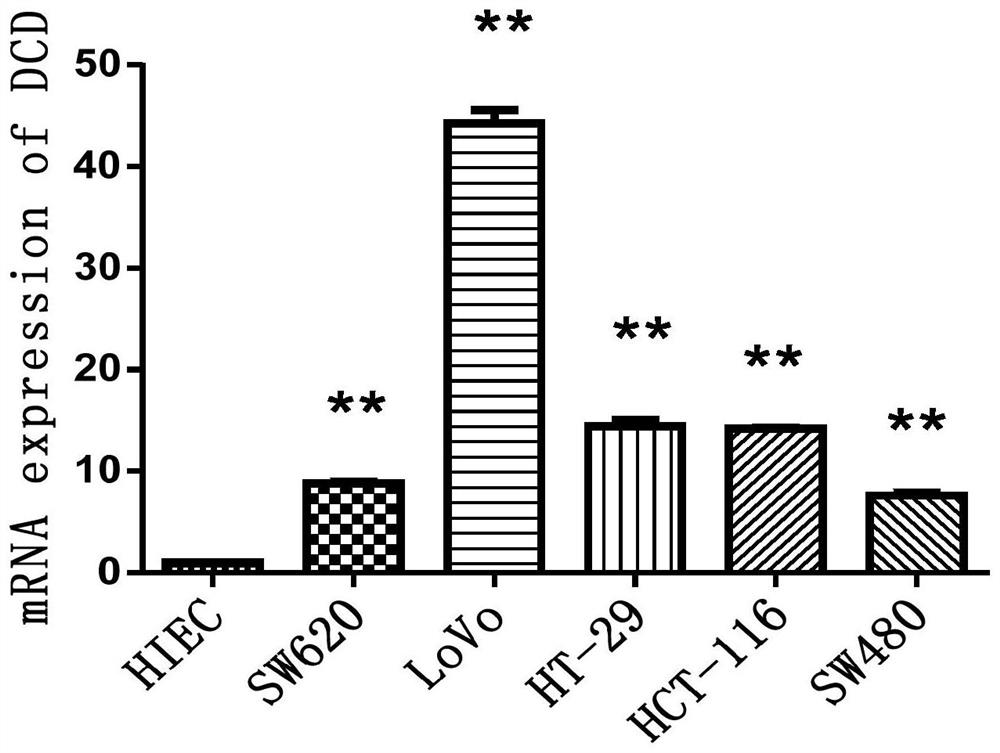
[0043] A Q-PCR detection method for colorectal cancer marker DCD, characterized in that it comprises the following steps: 1) selecting a normal human epithelial cell (HIEC) and five human colon cancer cells as cell samples; the five human colon cancer cells The cancer cells were SW620 cells, LoVo cells, HT-29 cells, HCT-116 cells and SW480 cells;
[0044]The RNA extraction method of the cell sample is as follows: After the cell sample has been resuscitated and cultured, add 1mL TRIzol RNA isolation reagent and let it stand at room temperature; then add 0.2mL chloroform, shake, and centrifuge at 4°C and 12000g for 15min; absorb the upper aqueous phase, transfer To another centrifuge tube, add 0.5mL isopropanol, mix well, let stand at 37°C for 10min, then centrifuge at 4°C, 12000g for 10min, discard the supernatant; continue to add 1mL 75% ethanol, shake Finally, centrifuge at 4°C and 7500g for 5min, discard the supernatant; finally dry naturally at 37°C, add 20 μL DEPC water to...
Embodiment 2
[0063] A Q-PCR detection method for colorectal cancer marker DCD, characterized in that it comprises the following steps:
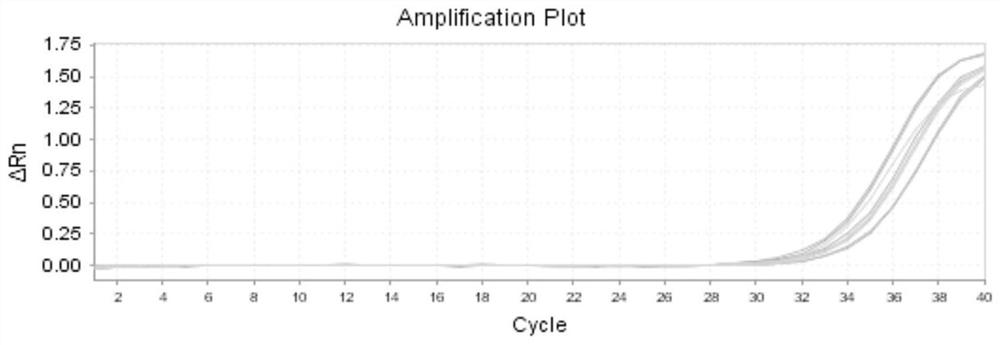
[0064] 1) Select five groups of colorectal cancer tissues (T) and five groups of paracancerous tissues (N is normal tissue) as tissue samples;
[0065] The RNA extraction method of tissue samples is as follows: cut 0.1g of tissue samples, add 1mL TRIzol RNA isolation reagent, and let it stand at room temperature; then add 0.2mL chloroform, after shaking, centrifuge at 4°C and 12000g for 15min; absorb the upper aqueous phase, transfer To another centrifuge tube, add 0.5mL isopropanol, mix well, let stand at 37°C for 10min, then centrifuge at 4°C, 12000g for 10min, discard the supernatant; continue to add 1mL 75% ethanol, shake Finally, centrifuge at 4°C and 7500g for 5min, discard the supernatant; finally dry naturally at 37°C, add 20 μL of DEPC water to obtain the RNA of the tissue sample.
[0066] 3) Reverse transcription: Prepare 20 μL of the first-str...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com