Method for constructing perilla frutescens core germplasm resource library based on SRAP molecular marker
A technology of core germplasm and molecular markers, applied in the field of genetic diversity evaluation and core germplasm construction, can solve the problems of increasing the difficulty of evaluating and utilizing germplasm resources, improper protection and utilization of perilla germplasm resources, and difficult to identify
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
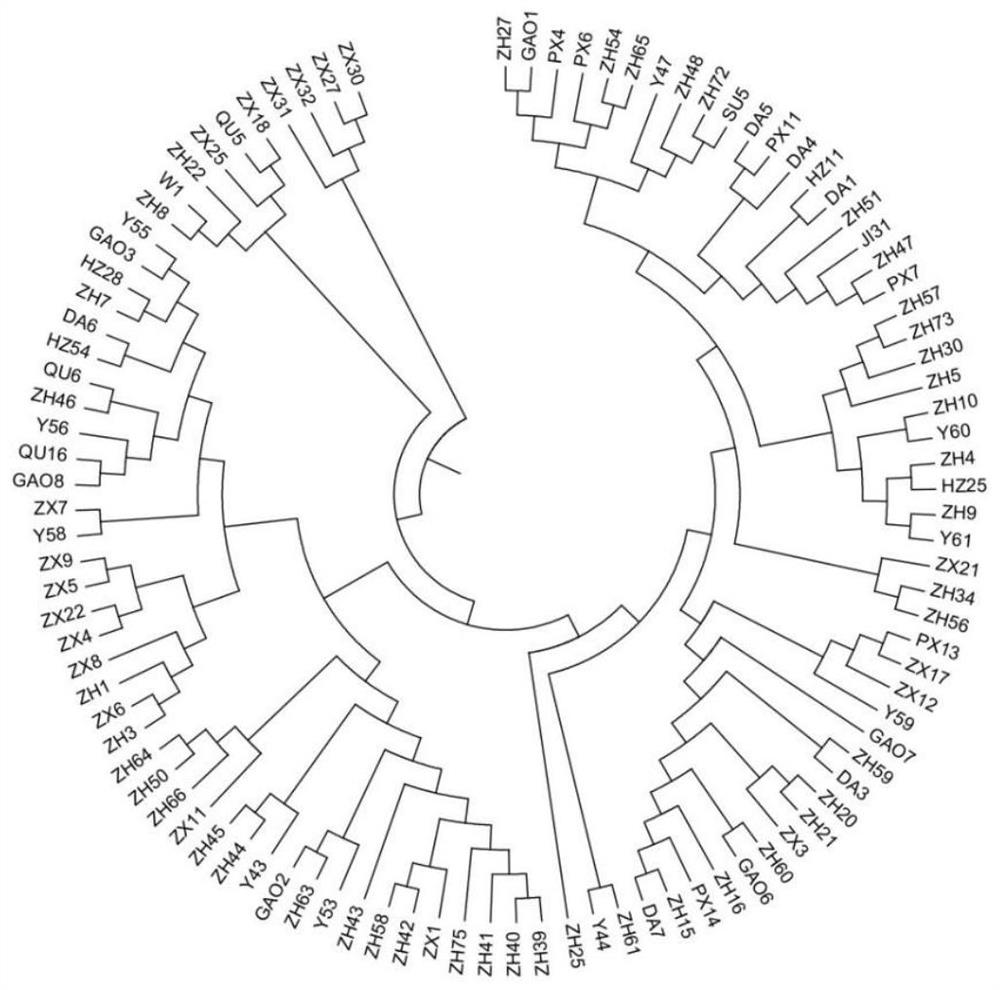
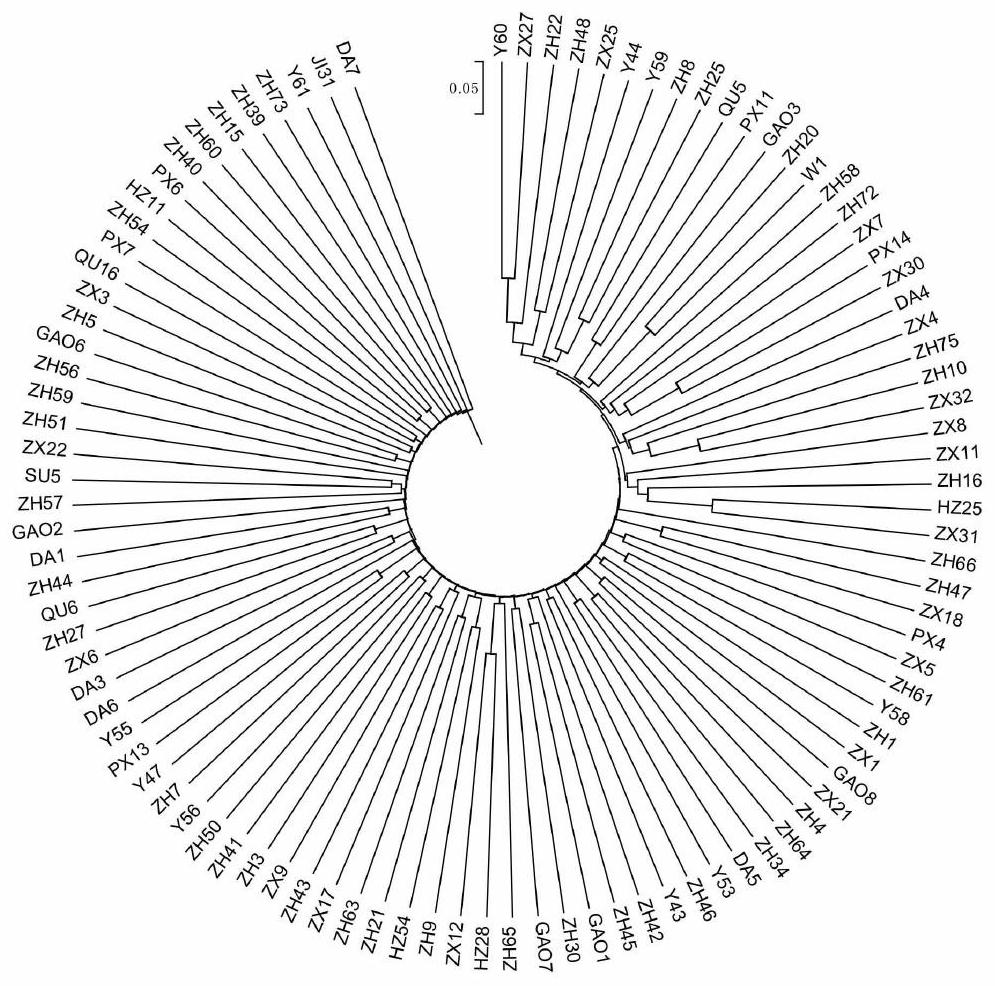
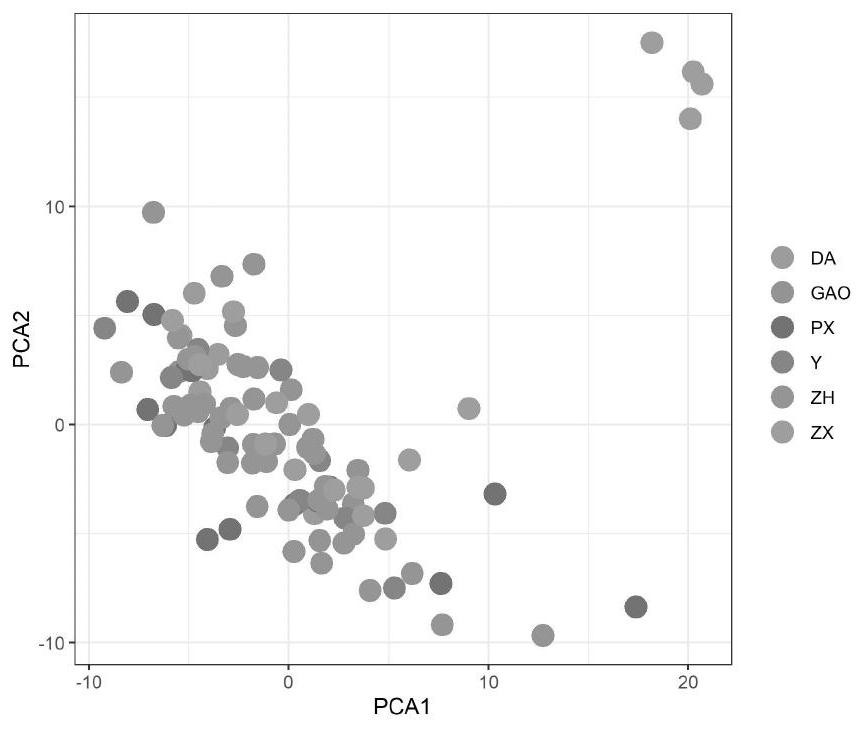
[0021] Specific embodiment one: what this embodiment records is a kind of method for constructing perilla core germplasm resource bank based on SRAP molecular marker, comprising the following steps:
[0022] Step 1: Through 8 years of field experiments, 100 perilla varieties were screened out, planted in flower pots, and the leaves of seedling plants were taken as test materials, the total DNA of each variety was extracted, and each variety was detected by 1% agarose gel electrophoresis. For the purity and integrity of the total DNA of each variety, use an ultraviolet spectrophotometer to measure the absorbance value of the total DNA of each variety at 260nm and 280nm to determine the purity and concentration of the DNA, and store it at -20°C for future use; after 8 years of field experiments, It is to prove that these seeds can be planted in Heilongjiang, and that they are classified according to their agronomic traits and have field stability;
[0023] Step 2: use the total ...
specific Embodiment approach 2
[0024] Specific embodiment 2: A method for constructing a perilla core germplasm resource bank based on SRAP molecular markers described in specific embodiment 1. In step 2, the forward primers used in the SRAP-PCR amplification are me1-me9 , the reverse primers are em1~em17. The SRAP amplification primers used are all existing ones, which can be obtained from Baidu.
[0025] The following are the specific primer design, debugging and amplification efficiency detection of the present invention. The sequences of primers and probes used in gene detection are as follows:
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com