Primer group for detecting non-coding small RNA by multiple fluorescent quantitative PCR based on stem-loop method
A multiplex fluorescence quantitative and fluorescent quantitative technology, applied in the field of fluorescence quantitative PCR, can solve the problems of complex components and complicated steps, and achieve the effects of preventing product contamination, easy operation and increasing accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 Primer and probe design
[0059] The design is mainly divided into 3 steps:
[0060] ① Design reverse transcription extension primers for the non-coding small RNA sequence to be tested.
[0061] ②Using the small RNA stem-loop reverse transcription method to design reverse transcription primers;
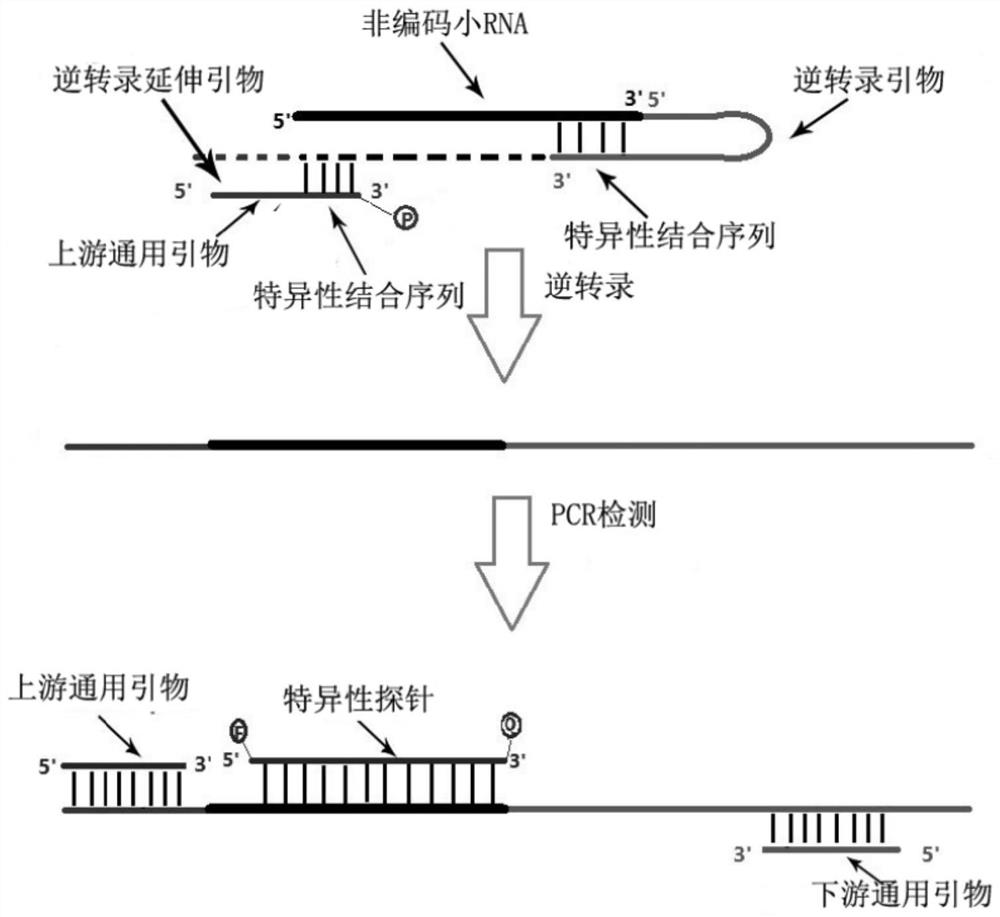
[0062] ③Design general upstream and downstream primers and specific probes corresponding to the corresponding sequences, and then detect non-coding small RNAs. The specific design ideas are as follows: figure 1 .
[0063] The specific design is as follows:
[0064] 1. Reverse transcription extension primer: the reverse transcription extension primer is sequenced from the 5' end to the 3' end: the upstream general primer sequence and the specific binding sequence 1; the specific binding sequence 1 is the same as the non-coding small The 6-15 bases of the 5' end of the RNA are the same; the sequence of the upstream universal primer is 16-25 bases; the 3' end of t...
Embodiment 2
[0069] Embodiment 2 Example of primer set design
[0070] Corresponding primers and probes were designed for miR-21-5p (nucleotide sequence shown in SEQ ID NO.1: UAGCUUAUCAGACUGAUGUUGA) by the method of Example 1.
[0071] As shown in Table 1, the 3' end of the designed reverse transcription extension primer has a length of 15 bases, and the underlined sequence is consistent with the 5' end sequence of the Has-miR-21-5p sequence (that is, specific binding or complementary sequence), the nucleotide sequence is shown in SEQ ID NO.2. At the same time, upstream universal primers, downstream universal primers, and specific probes are designed for the amplified fragments, and their nucleotide sequences are respectively shown in SEQ ID NO.3-6. The underlined sequence of the reverse transcription primer (i.e. RT primer) is the region complementary to the 3' end of the target sequence.
[0072] Table 1 Primer and probe sequence list
[0073]
Embodiment 3
[0074] Example 3 A detection method for detecting non-coding small RNAs by multiplex fluorescent quantitative PCR
[0075] 1. Extract sample non-coding small RNA
[0076] The miRcute kit kit was used to extract small non-coding RNA from samples, and the concentration and purity of RNA (OD260 / OD280) were determined by UV spectrophotometry.
[0077] 2. Reverse transcription
[0078] Use the reverse transcription extension primer and the stem-loop reverse transcription primer to perform reverse transcription, prepare the reverse transcription system in the PCR tube, then place the PCR tube in the ABI2720 PCR instrument, incubate at 25°C for 5min, at 50°C for 15min, and at 85°C for 5min, 4°C hold. The reversed product was immediately subjected to qPCR reaction or stored at -20°C.
[0079] Reverse transcription system: 5×RT buffer 4 μL; dNTPs (10 mM each) 1 μL; each RT extension primer (2 μM) of each non-coding small RNA to be tested 1 μL; each stem-loop reverse transcription pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com