Method for quantifying viable bacteria of flora and application of method
A technology of flora and live bacteria, applied in the field of microbial detection, can solve the problem of inaccurate distinction between dead and live bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0024] Preparation of the flora
[0025] Dissolve 100-200g of feces collected from healthy donors in 750mL-1L of 0.9% NaCl solution, and process them through the feces analysis pretreatment instrument TG-01 (patent number CN201930312740.2) and supporting consumables produced by Chengge Biotechnology Co., Ltd. Remove the residue to obtain 30-40g of fecal bacteria;
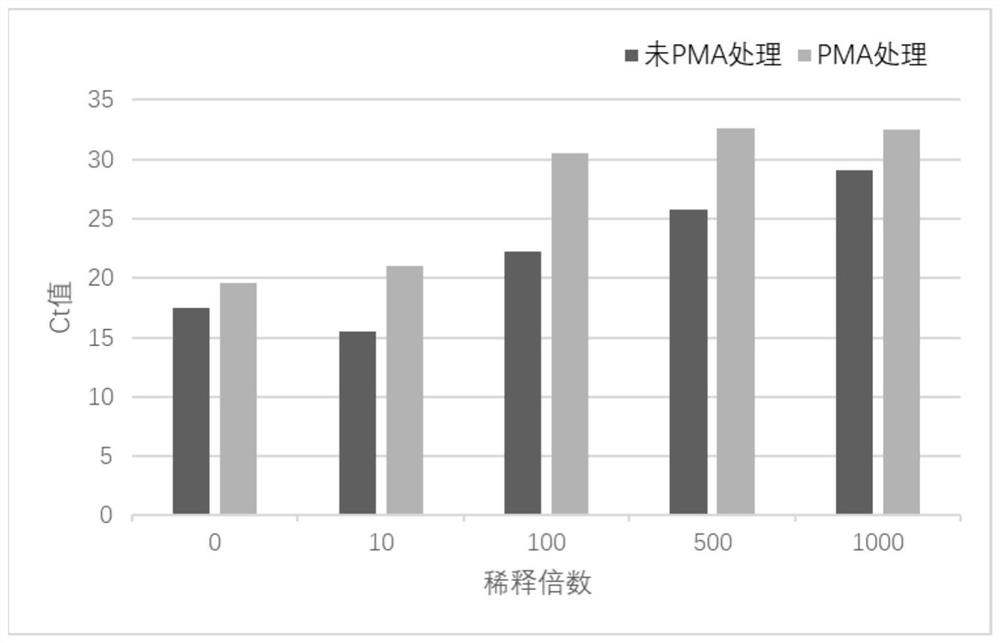
[0026] Dilute 10 times, 100 times, 500 times, 1000 times with PBS buffer containing 0.05% L-cysteine hydrochloride, and sterilize at 115°C for 10 minutes to prepare heat-inactivated fecal bacteria suspension, respectively Pipette 500 μL into a 1.5ml centrifuge tube, perform PMA treatment according to step 1.2.1, and use no PMA treatment as a control, then use the kit method (QIAamp Fast DNAStool Mini Kit) to extract genomic DNA, perform qPCR detection, and use the Ct value as indicators to analyze the results.
[0027] See the experimental results figure 1 ,Depend on figure 1 It can be seen that in the undilut...
Embodiment 1
[0034] The live bacteria quantitative method of flora comprises the following steps:
[0035] Prepare the flora, dissolve the collected samples in 0.9% NaCl solution, and remove the residue through the stool analysis pretreatment instrument TG-01 (patent number CN201930312740.2) produced by Chengge Biotechnology Co., Ltd. and supporting consumables to obtain the flora , samples include feces, soil, silt, etc.;
[0036] Preparation of PMA stock solution: Dissolve PMA in ddH 2 O, the PMA mother solution with a solubility of 2mM was obtained, and the PMA mother solution was stored in the dark at -20°C;
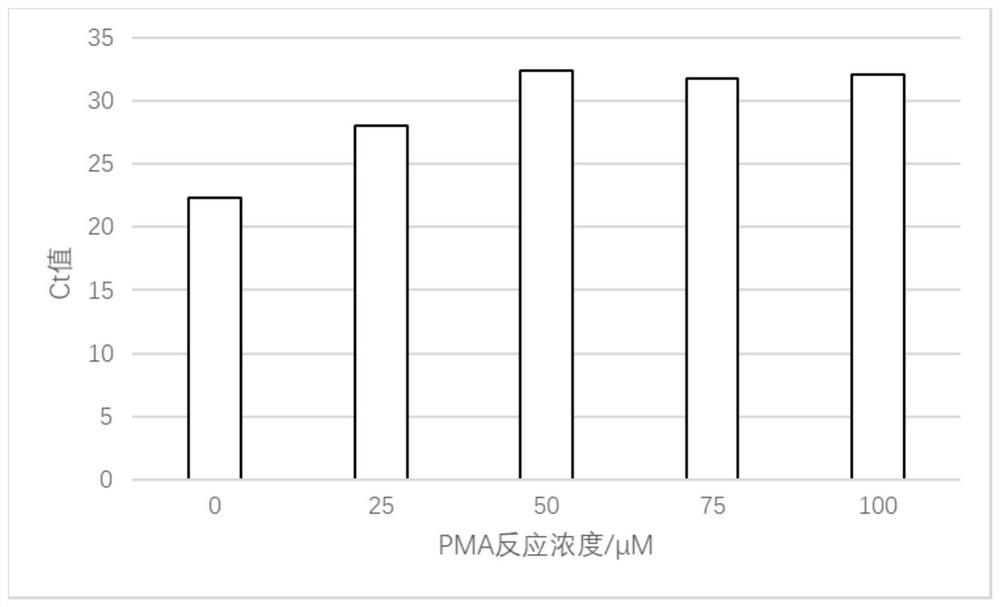
[0037] Preparation of PMA bacterial suspension: Take part of the bacterial population, dilute it 100 times with PBS buffer containing 0.05% L-cysteine hydrochloride, add PMA to make the final concentration of the system 50 μM, and mix well to obtain the PMA bacterial suspension ;
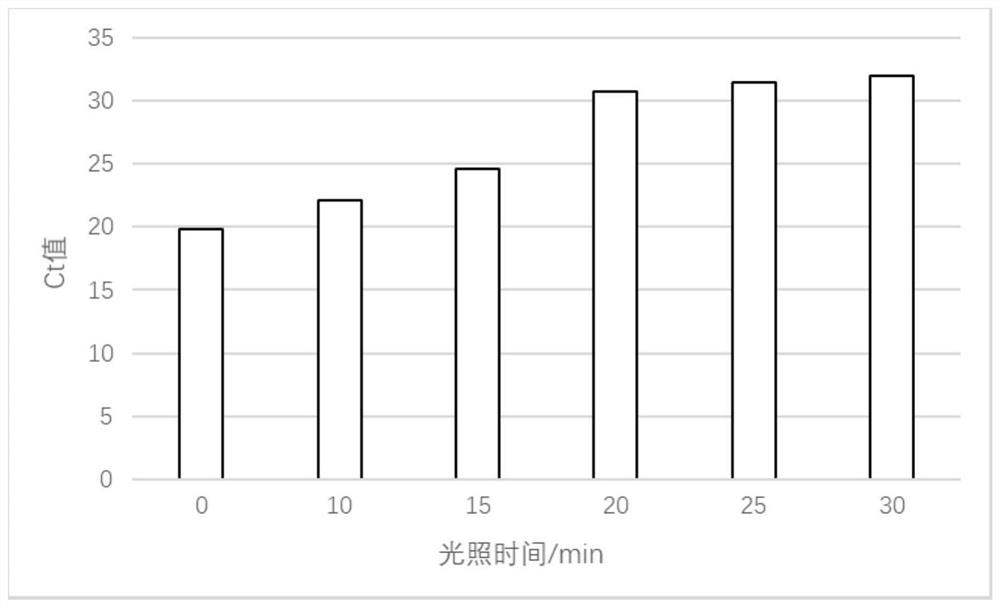
[0038]Treatment of PMA bacterial suspension: incubate the obtained PMA bacterial suspension at...
Embodiment 2
[0040] Use the excrement bacterium that embodiment 1 obtains;
[0041] Preparation of PMA stock solution: Dissolve PMA in ddH 2 O, the PMA mother solution with a solubility of 2mM was obtained, and the PMA mother solution was stored in the dark at -20°C;
[0042] Preparation of PMA bacterial suspension: Take part of the bacterial population, dilute it 100 times with PBS buffer containing 0.05% L-cysteine hydrochloride, add PMA to make the final concentration of the system 50 μM, and mix well to obtain the PMA bacterial suspension ;
[0043] PMA bacterial suspension treatment: incubate the obtained PMA bacterial suspension at room temperature in the dark for 30 minutes, then place the sample on ice, use a 500w halogen lamp to illuminate for 20 minutes at a distance of 20cm, and rotate at 5000r / min at 4°C after illumination Centrifuge for 3-5min, collect the cells for DNA extraction, and perform qPCR detection and analysis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com