Characterization and application of Novel high-temperature Argonaute protein TpsAgo
A reaction and reaction system technology, applied in the biological field, can solve problems such as the scarcity of circulating tumor DNA, sensitivity, and operating cost limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0226] Embodiment 1: Obtaining of TpsAgo gene sequence
[0227] In the database, the amino acid sequence of known PfAgo was searched for similarity, some amino acid sequences with high sequence consistency were selected, analyzed by MEGA software, a homologous evolution tree was constructed, and TpsAgo was selected as a candidate enzyme. The amino acid sequence of TpsAgo (WP_060384876.1) and the corresponding gene sequence (NZ_CP014142.1) encoding the protein were obtained. After the gene sequence was synthesized by codon optimization, it was cloned into pET28a expression vector.
Embodiment 2
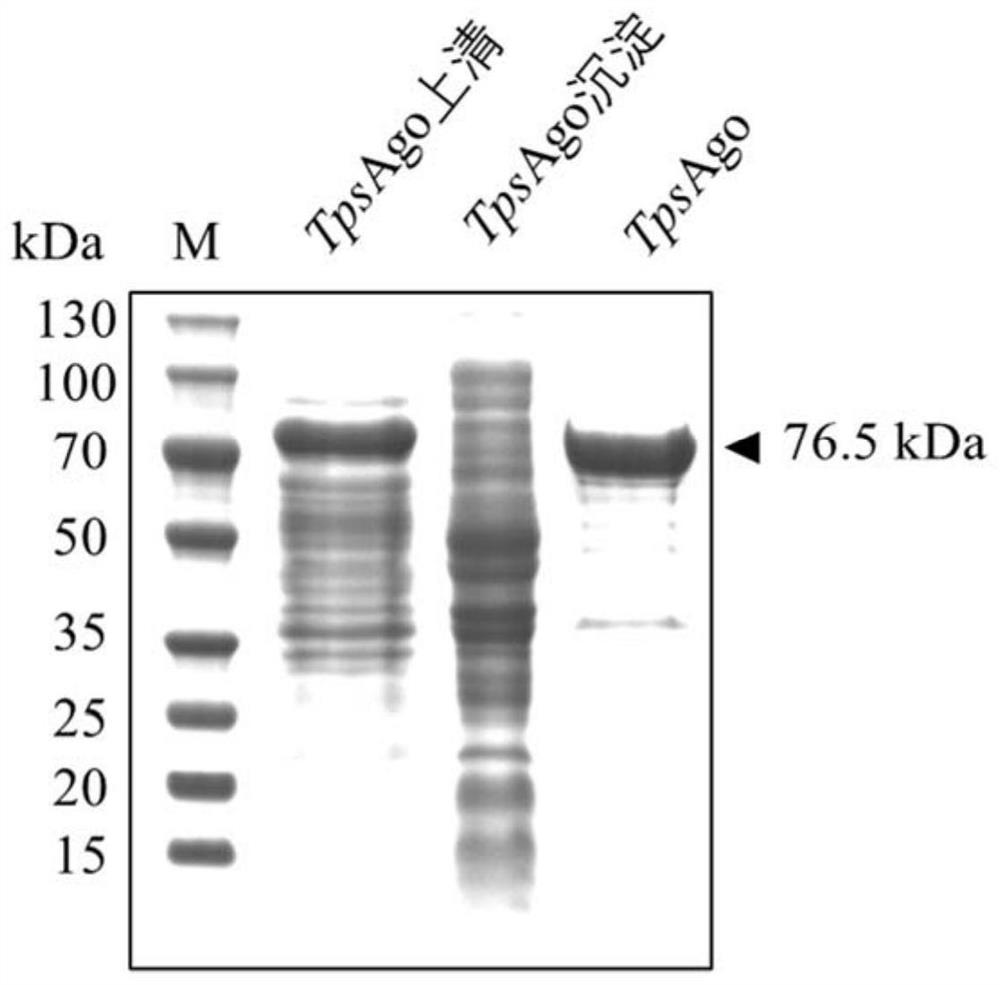
[0228] Example 2: Heterologous expression and purification of TpsAgo protein
[0229] The above TpsAgo-pET28a prokaryotic expression plasmid was introduced into E.coli BL21(DE3) to obtain the TpsAgo-pET28a / E.coli BL21(DE3) prokaryotic expression strain. The expression strain E.coliBL21(DE3) containing the recombinant plasmid TpsAgo-pET28a was inoculated in LB medium containing 50 μg / mL kanamycin, and cultivated to OD at 37°C and 220rpm on a shaker 600 Between 0.6-0.8, add IPTG with a final concentration of 0.4-0.6mM, continue to culture at 18°C, 200rpm shaker for 16-20h, and induce the expression of TpsAgo protein. Collect the cells by centrifugation, resuspend the cells in a resuspension buffer (containing 20mM Tris-HCl, pH around 8.0, 1M NaCl), then crush the cells by high pressure, and centrifuge to obtain the supernatant. Using Ni-NTA column to affinity purify the protein, the eluate is concentrated by ultrafiltration, desalted and other steps to obtain the purified prote...
Embodiment 3
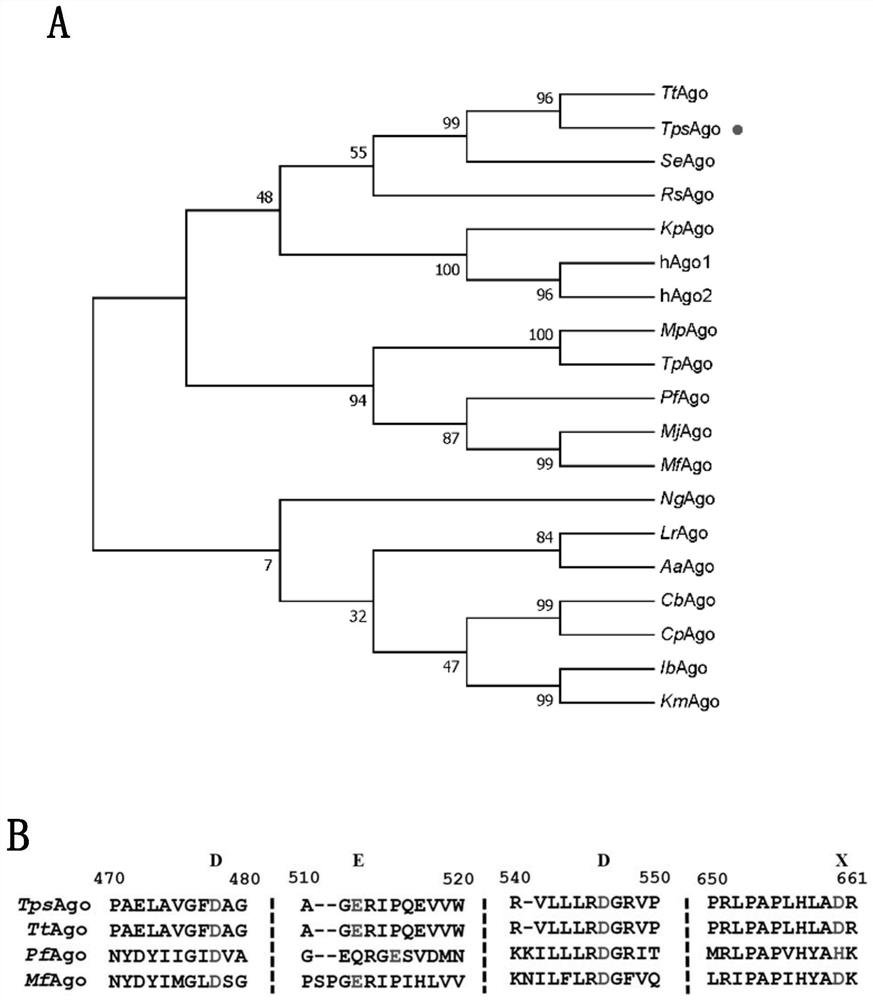
[0231] Example 3: Alignment of TpsAgo with other known Ago sequences
[0232] In this example, multiple sequence alignments were performed between TpsAgo and partially characterized Ago.
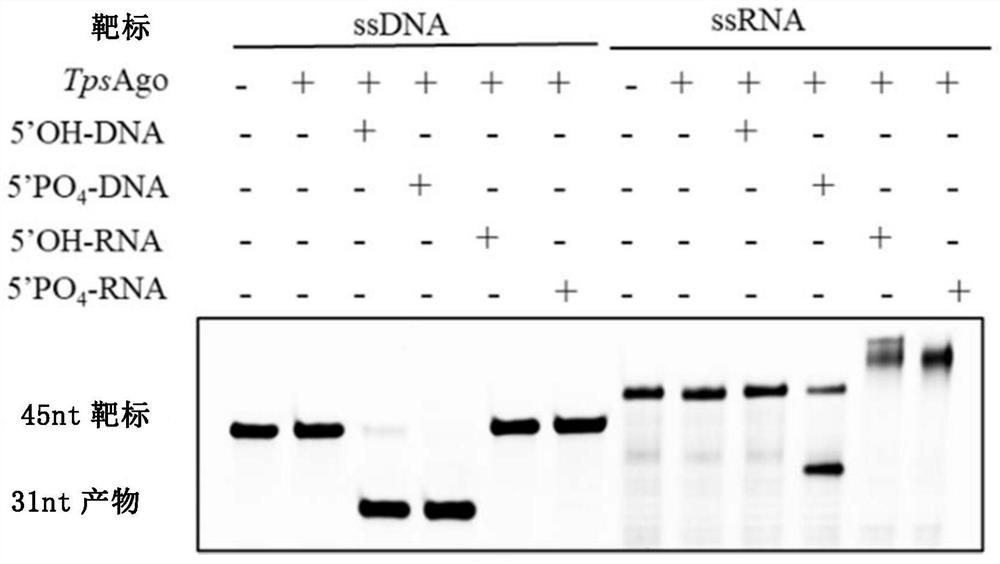
[0233] The results showed that TpsAgo had the least number of amino acids and the smallest protein molecular weight. According to literature reports, the targeted cleavage of all catalytically active Ago proteins is mediated by a conserved DEDX (X stands for histidine, aspartic acid or asparagine) quadruple. Through sequence comparison, it can be found that there is a DEDD quadruple in TpsAgo ( figure 1 ). Therefore, it is further speculated that it may have nuclease catalytic activity, which requires further identification and characterization in vitro.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com