Rapid feature selection method based on whole genome sequence SNP data
A feature selection method, a genome-wide technology, applied in genomics, proteomics, instruments, etc., can solve problems such as low accuracy, low efficiency of feature selection for high-dimensional samples, limited improvement in analysis efficiency and accuracy, and achieve Improve the accuracy, improve the efficiency of SNP feature selection, and improve the effect of credibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
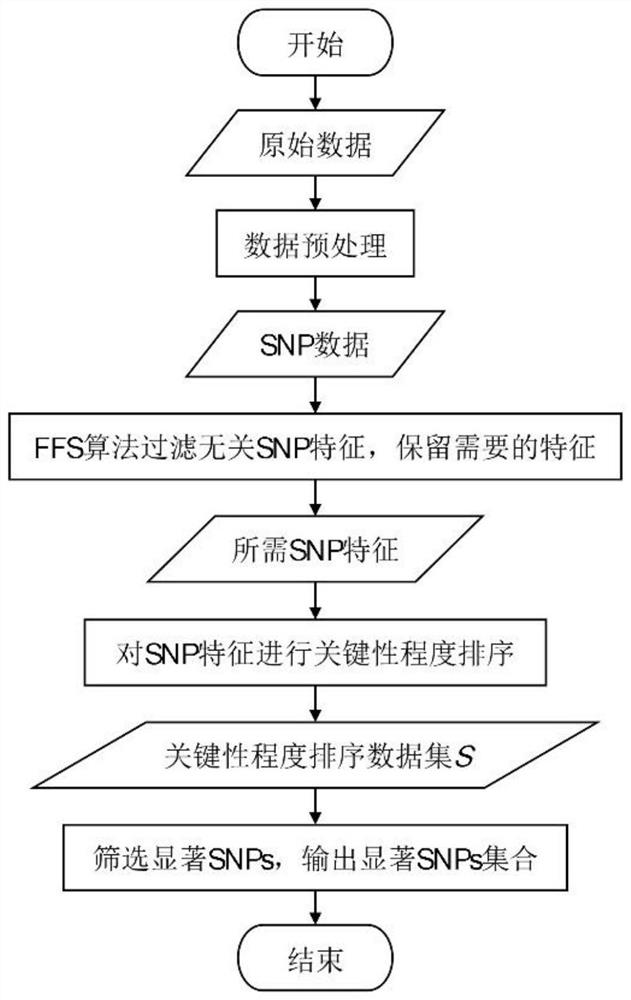
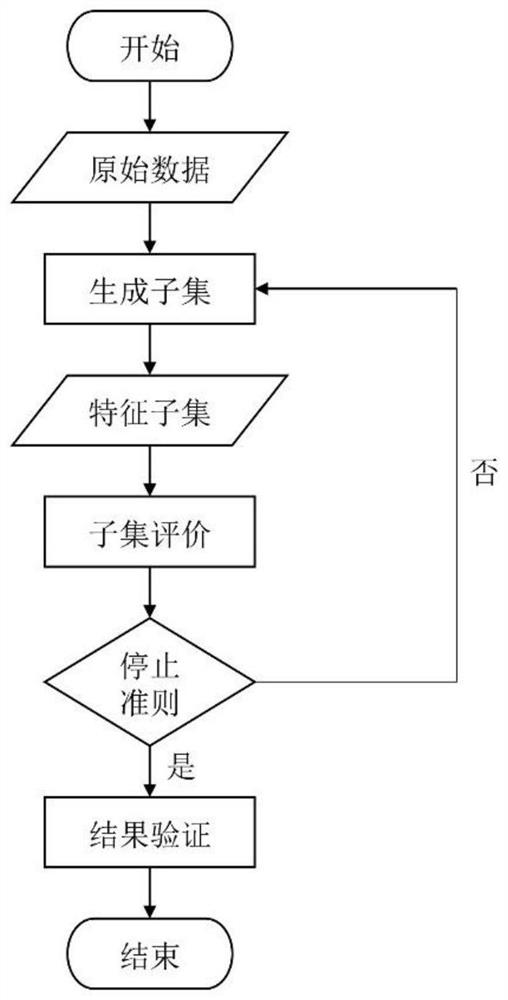
[0045] A fast feature selection method based on whole-genome sequence SNP data to figure 2The process shown is the basic framework, and the specific process is as follows figure 1 shown, including the following steps:
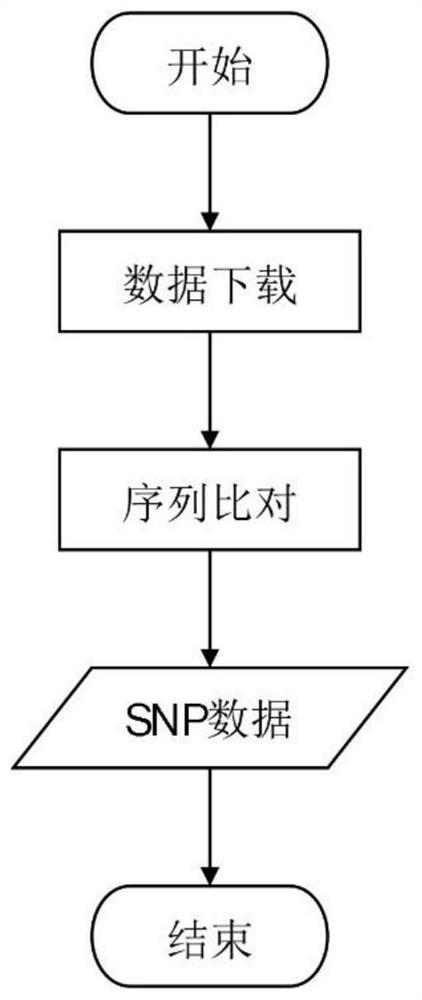
[0046] (A) Preprocess the whole genome sequence data to obtain all SNP sites, the specific process is as follows image 3 shown, including the following steps:
[0047] A1) Obtain reference sequences and test sequences from databases;
[0048] A2) Use MUMmer 3 software to compare the reference sequence and the test sequence to obtain all SNP sites.
[0049] (B) Use the FFS algorithm to filter irrelevant SNP features and retain the required SNP features. The specific process is as follows: Figure 4 shown, including the following steps:
[0050] Use all SNP sites obtained in step (A) as string Enter a regular expression re. findall (pattern, string, flags=0) In, output the required SNP features.
[0051] (C) Sorting the criticality of the SNP featu...
Embodiment 2
[0057] A fast feature selection method based on whole genome sequence SNP data, comprising the following steps:
[0058] (A) Preprocess the whole genome sequence data to obtain all SNP sites, the specific process is as follows:
[0059] A1) Obtain reference sequences and test sequences from databases;
[0060] A2) Use MUMmer 3 software to compare the reference sequence and the test sequence to obtain all SNP sites.
[0061] (B) Use the FFS algorithm to filter irrelevant SNP features and retain the required SNP features. The specific process is as follows:
[0062] Use all SNP sites obtained in step (A) as string Enter a regular expression re. findall (pattern, string, flags=0) In, output the required SNP features.
[0063] (C) Sorting the criticality of the SNP features retained in step (B), the specific process is as follows:
[0064] Add the SNP features retained in step (B) to the criticality ranking data set S In the data set S as iterable input expression So...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com