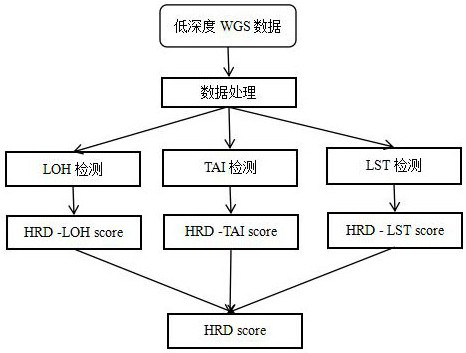
A method for evaluating hrd score based on low-depth wgs
A deep, computationally based technology, applied in the fields of genomics, instrumentation, sequence analysis, etc., can solve the problems of high cost and high accuracy, and achieve the effect of reducing costs and facilitating large-scale applications.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] Example 1 Preprocessing of low-depth WGS data processing
[0089] 1. Use the fastp software to remove the joints on the reads from the low-depth WGS off-machine data;
[0090] 2. Compare the processed off-machine data with the reference genome of the whole human genome through bwa software, and obtain the first comparison file in bam format;
[0091] 3. Correct the base quality value of the first comparison file through GATK software;
[0092] 4. Use the picard software to remove duplicate reads from the corrected first alignment file to obtain a second alignment file that does not contain duplicate reads. The format of the file is bam format.
[0093] 5. Divide the whole human genome into windows of 100Kbp size according to the order of arrangement.
Embodiment 2
[0094] Example 2 Construction of DR fragments
[0095] 1) Taking the reads in the second comparison file in Example 1 as the basic unit, count the number of reads falling in each window in Example 1, as the reads count of the window, and record it as RC i , i is the order of the windows divided in the whole genome according to the order of arrangement, and i is 1, 2, 3....
[0096] 2) Count the GC base content of each window, merge adjacent windows with the same GC content into one group, and record the jth group as W j , the number of windows contained in group j is denoted as M j, the kth window contained in the jth group is recorded as W kj , j, k are 1,2,3....;
[0097] 3) Calculate W j The median value RC of , denoted as RC j , and the average RC of the sample to be tested as a whole, denoted as RC p , by the following formula for RC i Make corrections:
[0098]
[0099] i=M 1 +M 2 +M 3 ...+M (j-1) +k;
[0100] 4) Process the low-depth WGS data of 30 healt...
Embodiment 3
[0103] Example 3 Calculation of copy number
[0104] Count the median value of DR in each DR segment, as the DR value of the DR segment, recorded as DR q , calculate the copy number of the DR segment, denoted as C q , the calculation formula is:
[0105] .
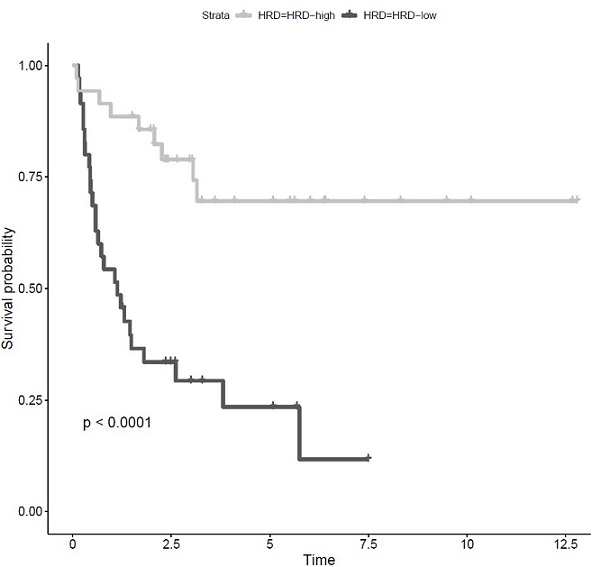
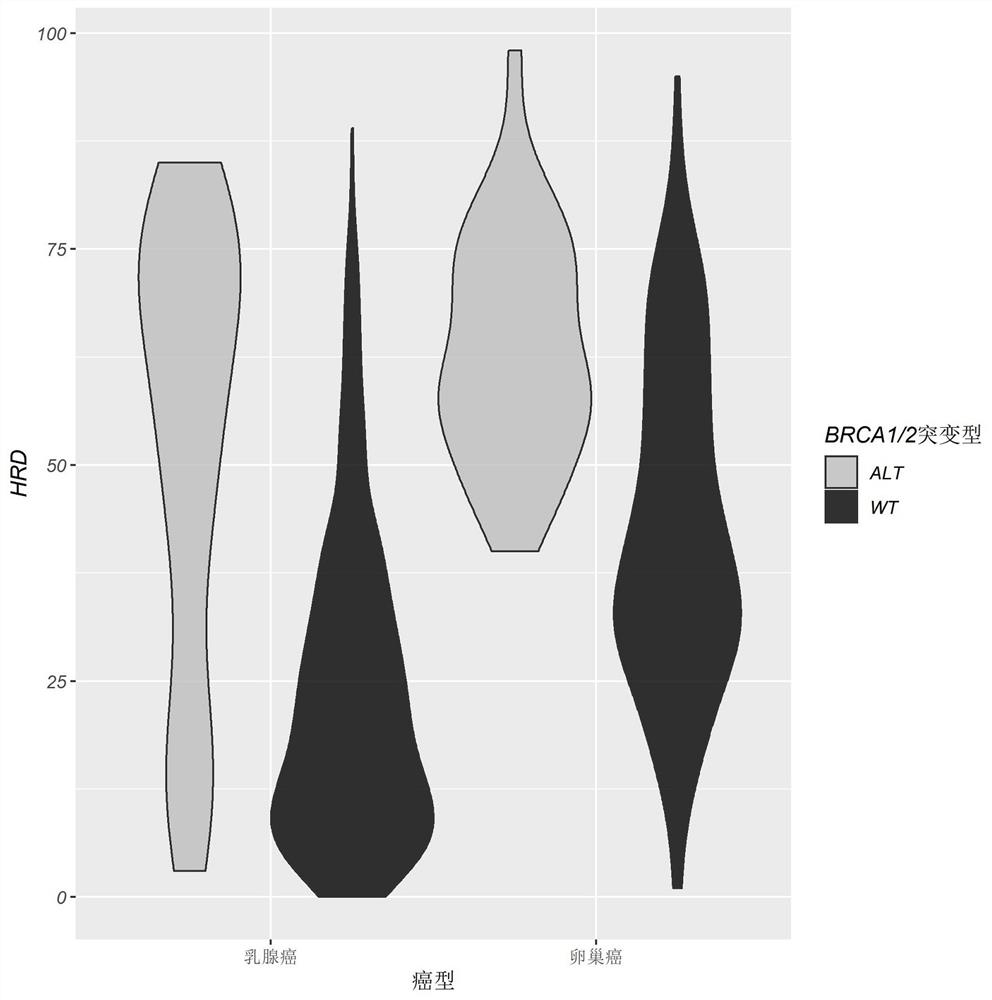
[0106] In this embodiment, the internal cause of the patient's cancer can be preliminarily understood through the calculation of Cq (copy number).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com