Method and device for detecting exon level deletion of target gene based on reads depth
An exonic, in-depth technology, applied in genomics, instrumentation, sequence analysis, etc., can solve the problem of unable to detect the type of copy number chromosomal mutation, and achieve the effect of clear results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
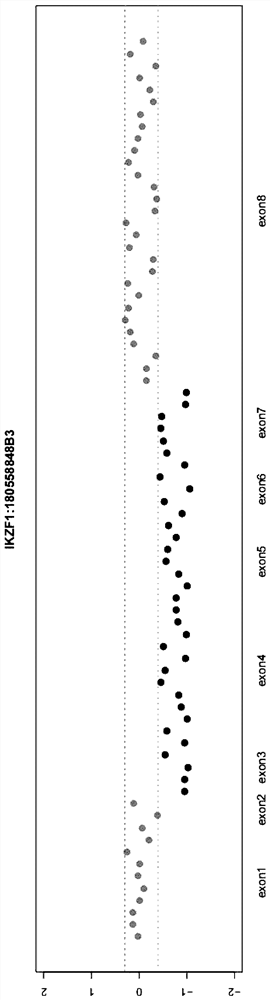
[0056] OBJECTIVE: To detect IKZF1 deletion status in patient samples of hematological diseases, especially children.
[0057] step:
[0058] 1. For 30 patients with hematological diseases who did not submit paired samples, the IKZF1 deletion status was detected.
[0059] 2. Use the intermediate output file generated by the algorithm detection of this paper as input for visual display.
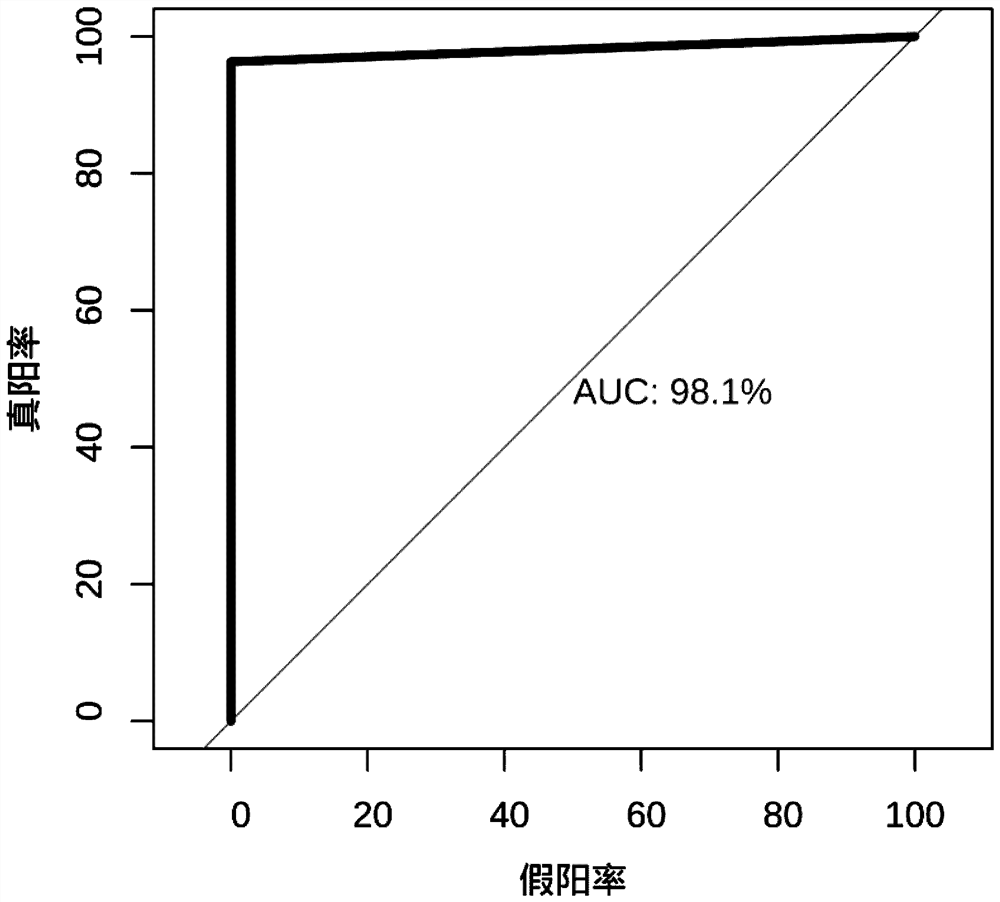
[0060] 3. Experimental verification of MLPA technology was carried out on these 30 patients.
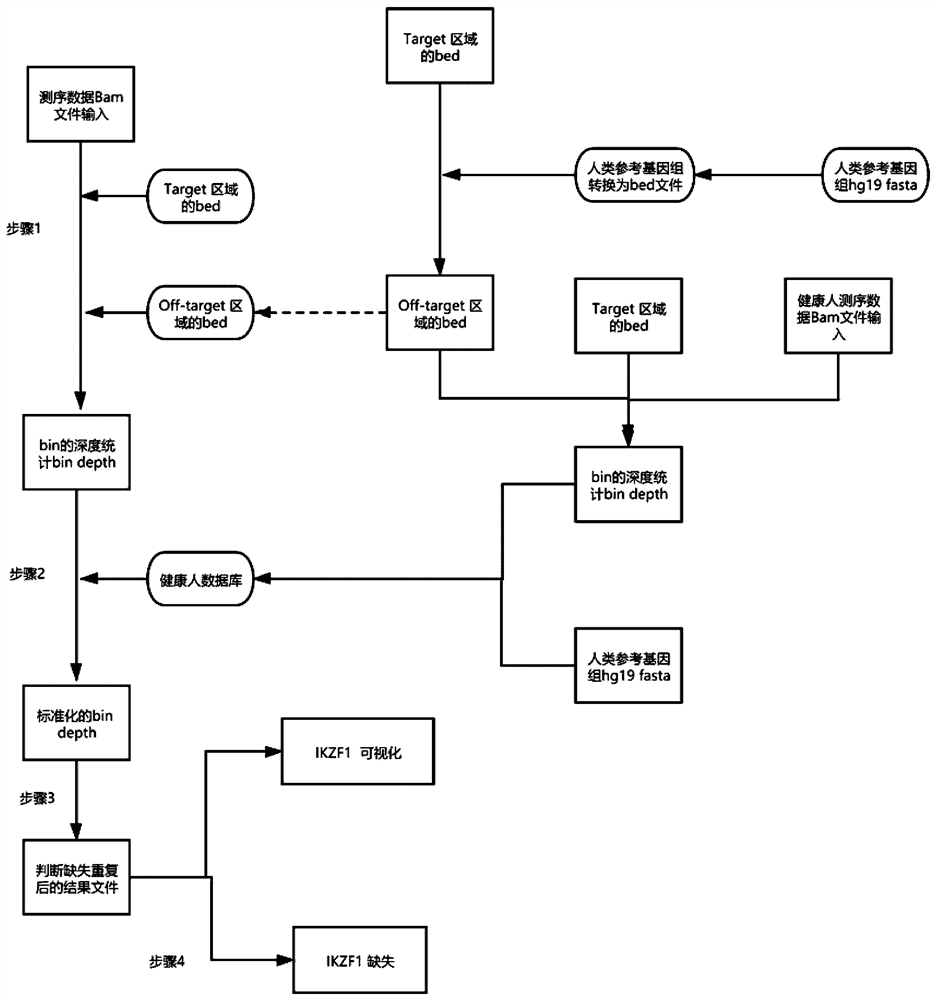
[0061] Among them, the detection of IKZF1 missing state mainly includes the following steps:
[0062] First, the first step is to divide the genome into multiple bins, which are divided into bins in the target area and bins in the off-target area, compare the reads to the reference genome, and calculate each The log2 value of the average read depth and depth in the bin;
[0063]Second, the read depth statistics of the target region and the off-target region are merged and normalized. In a typical...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com