Construction Method of Peanut Genetic Population
A technology for genetic populations and construction methods, used in biochemical equipment and methods, botanical equipment and methods, and microbial assay/inspection, etc., and can solve problems such as low polymorphism, low efficiency, and cumbersome screening work.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
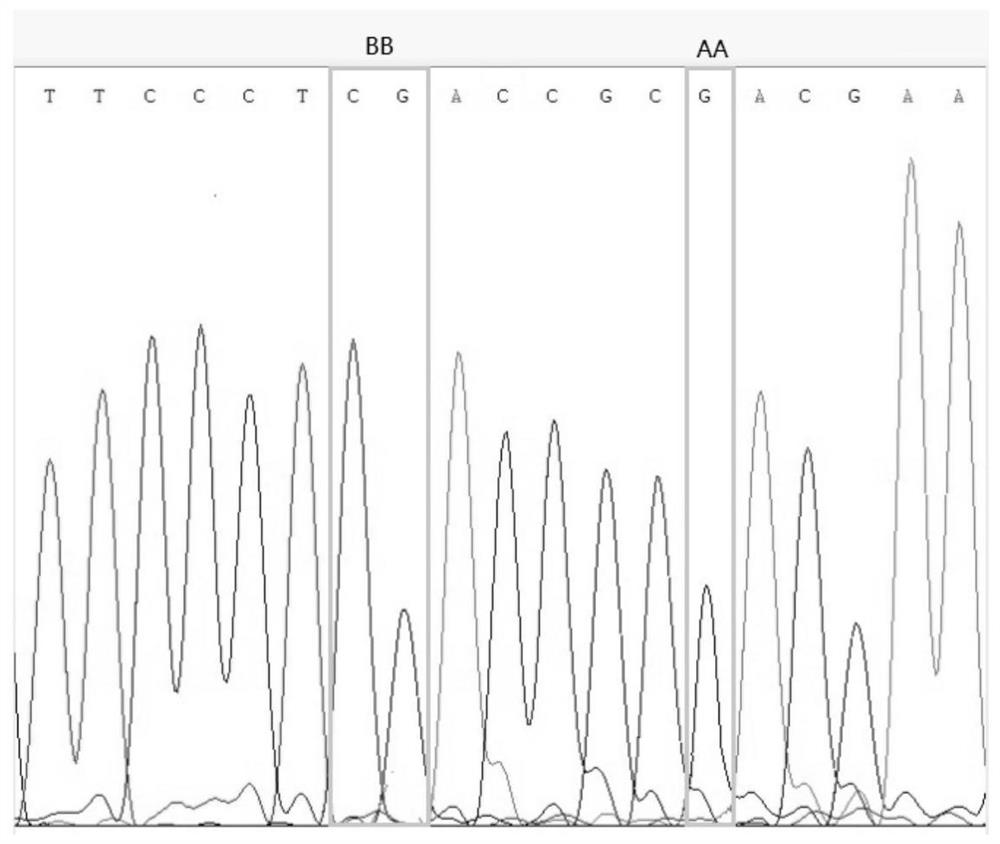
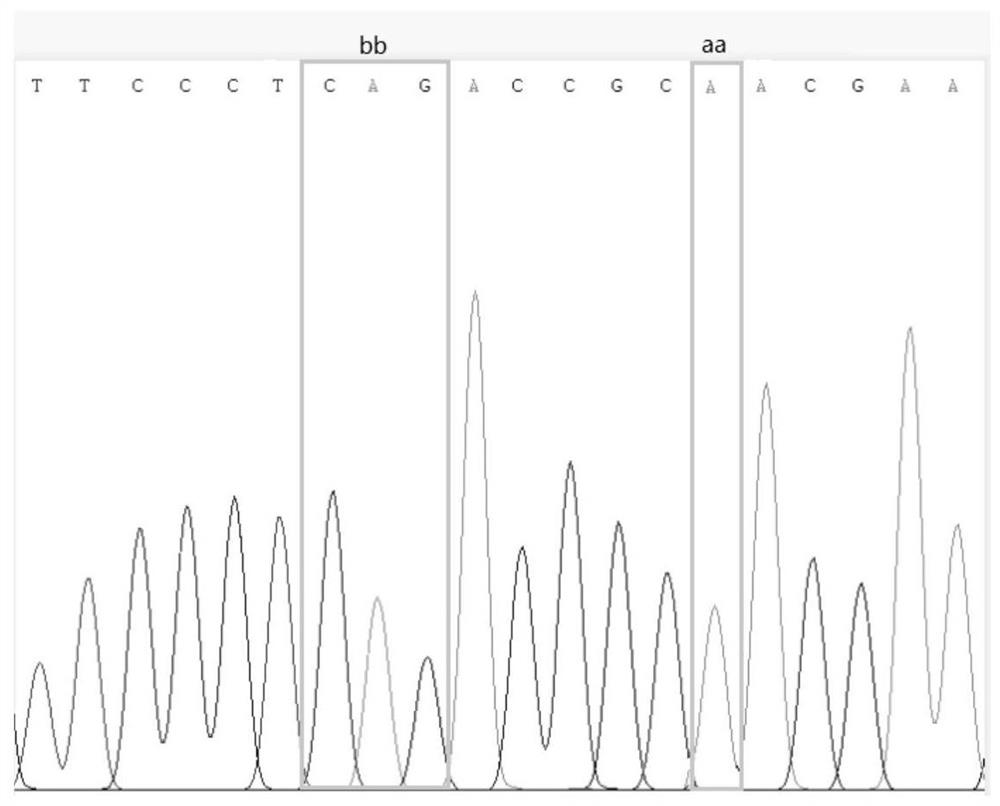
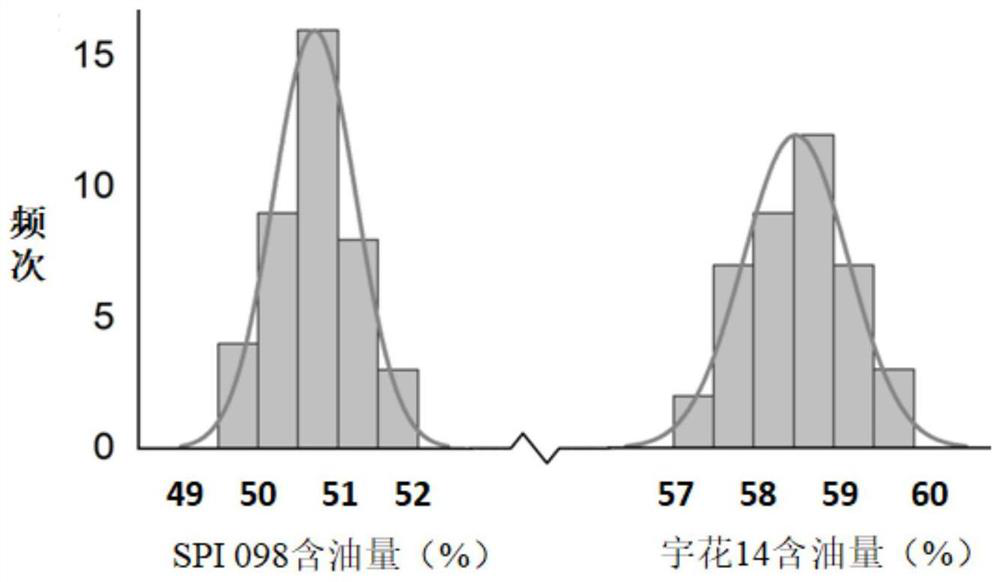
[0045] Parents: Peanut high-oil parent Yuhua 14, FAD2 loci are AhFAD2A and AhFAD2B (genotype is AABB); peanut low-oil parent SPI098, FAD2 loci are AhFAD2a and AhFAD2b (genotype is aabb), both FAD2 loci genotype differences such as figure 1 and figure 2 As shown, there is a significant difference in oil content between Yuhua 14 and SPI098, as shown in image 3 shown.
[0046] The construction method of peanut genetic population, the steps are as follows:
[0047] 1. Artificial pollination hybridization of peanuts
[0048] The high-oil parent Yuhua 14 (common oleic acid content) and the low-oil parent SPI098 (high oleic acid content) were planted at intervals in the field. One week before the full flowering period (about 7 weeks after sowing), the flower buds of the female parent Yuhua 14 were manually removed for about 1 week; the anthers in the female parent flower buds were removed with tweezers after 17:00 every afternoon in the full flowering period, and the detasselin...
Embodiment 2
[0068] Parents: large peanut variety Luhua 11, FAD2 loci of AhFAD2a and AhFAD2B (genotype aaBB), peanut small fruit line BC54 crossed between Luhua 11 and Kainong 1715, then backcrossed with female parent Luhua 114 The FAD2 locus is AhFAD2a and AhFAD2b (genotype is aabb), and the genotype difference of the two FAD2 loci is as follows Figure 7 and figure 2 As shown, there is a significant difference in pod length between Luhua 11 and BC54, such as Figure 8 shown.
[0069] A method for constructing a peanut genetic population, the steps are as follows:
[0070] 1. Artificial pollination hybridization of peanuts
[0071] The large-fruit parent Luhua 11 (common oleic acid content) and the small-fruit parent Luhua 11 and Kainong 1715 backcross progeny BC54 (high oleic acid content) were planted at intervals in the field. One week before the full flowering period (about 7 weeks after sowing), the flower buds of the female parent Luhua 11 were manually removed for about one we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com