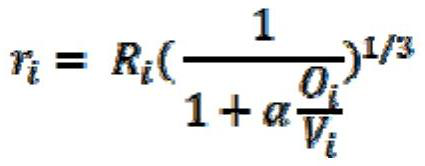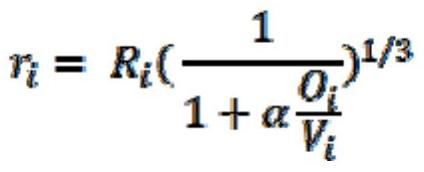Molecular volume calculation method based on variable radius Gaussian ball and molecular three-dimensional similarity scoring method for virtual drug screening
A technology of molecular volume and calculation method, which is applied in the field of computer-aided drug research and development, can solve the problems of no elimination error, poor accuracy, high false positive or false negative in drug virtual screening, etc., to achieve accuracy improvement, increase precision, and maintain calculation speed Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1: Molecular volume calculation method based on variable radius Gaussian sphere
[0042] The molecular volume calculation method based on the variable radius Gaussian sphere of the present embodiment comprises the following steps:
[0043] Read in the three-dimensional structure information of the molecule, the three-dimensional structure information includes the type and coordinate value of each atom in the molecule.
[0044] According to the type of each atom in the molecule, the van der Waals radius of the atom is obtained, and the three-dimensional structure information is converted into a set of Gaussian spheres representing each atom in the molecule, that is, each atom is represented by a Gaussian sphere, and the radius of each Gaussian sphere is not equal to that of the atom. Van der Waals radius, but smaller than its van der Waals radius, the formula for calculating the adjusted Gaussian sphere radius is as follows:
[0045]
[0046] Among them: f...
Embodiment 2
[0049] Example 2: A molecular three-dimensional similarity scoring method for virtual drug screening
[0050] In this embodiment, the target ADA17 in the DUD-E data set is taken as an example. There are 102 biological target information in the DUD-E data set, and each target has a corresponding active molecule set and a Decoy molecule set. Among them, the ADA17 target data set contains 1,341 active molecule sets and 35,900 Decoy molecule sets. The active molecule in the crystal structure is selected as the template molecule (hereinafter referred to as "molecule A") and the first molecule in the set of active molecules (hereinafter referred to as "molecule B") respectively.
specific Embodiment approach
[0051] A specific embodiment of a molecular three-dimensional similarity scoring method for virtual drug screening, comprising the following steps:
[0052] Step 1. Molecular volume calculation method based on variable radius Gaussian sphere:
[0053] This step is exactly the same as that of Example 1.
[0054] Step 2. Based on the variable radius Gaussian sphere, calculate various characteristic parameters of the two molecules used for similarity comparison:
[0055] Read the topological structure and three-dimensional structure information of molecule A and molecule B. The three-dimensional structure information includes the total number of atoms in the molecule, the total number of chemical bonds, the type of each atom and its coordinate value, and then calculate the similarity according to the following steps. Feature parameters for comparison:
[0056] Step 1. Take the absolute value of the difference between the total number of atoms of molecules A and B to obtain the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com