Method for recognizing antioxidant protein based on interaction of amino acid composition and protein
An antioxidant protein and amino acid technology, applied in the field of medicine, can solve the problems of low accuracy of antioxidant protein identification, protein secondary structure error, etc., and achieve the effects of increasing model identification efficiency, improving work efficiency, and optimizing the identification effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
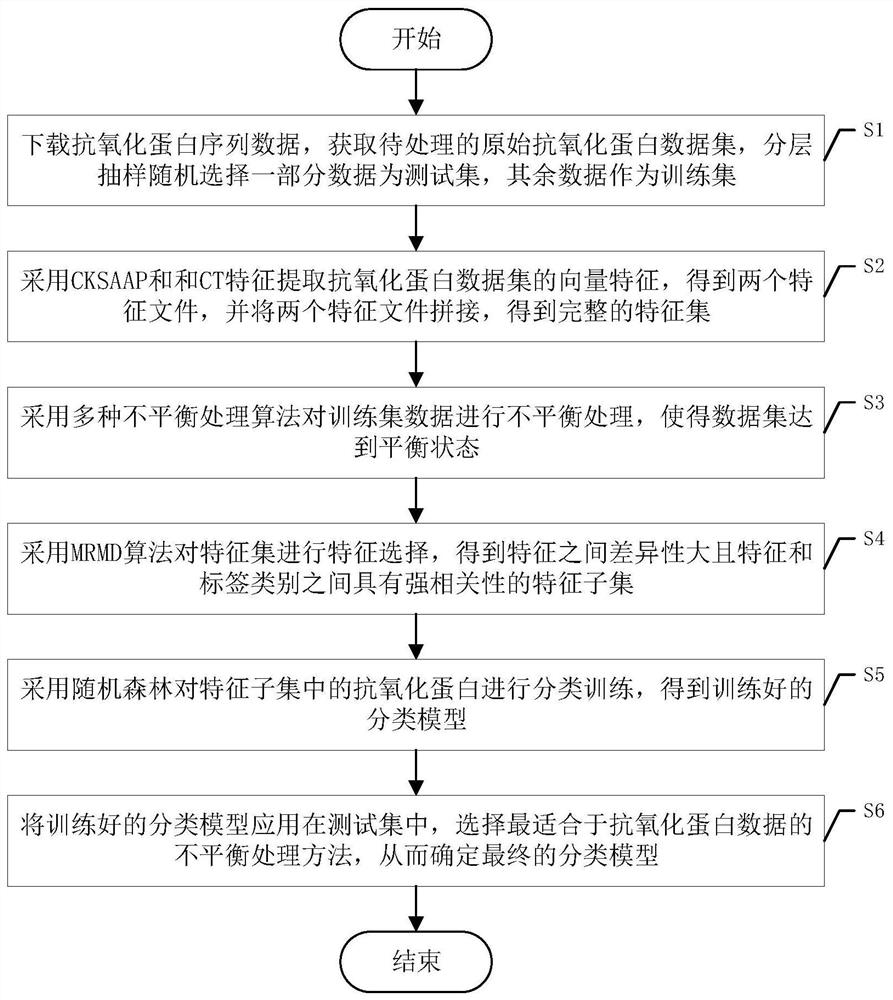
[0050] Specific implementation mode one: refer to figure 1 Specifically explaining this embodiment, the method for identifying antioxidant proteins based on amino acid composition and protein interaction described in this embodiment includes the following steps:
[0051] S1. Download the antioxidant protein sequence data, obtain the original antioxidant protein data set to be processed, randomly select a part of the data as the test set by stratified sampling, and use the rest of the data as the training set.
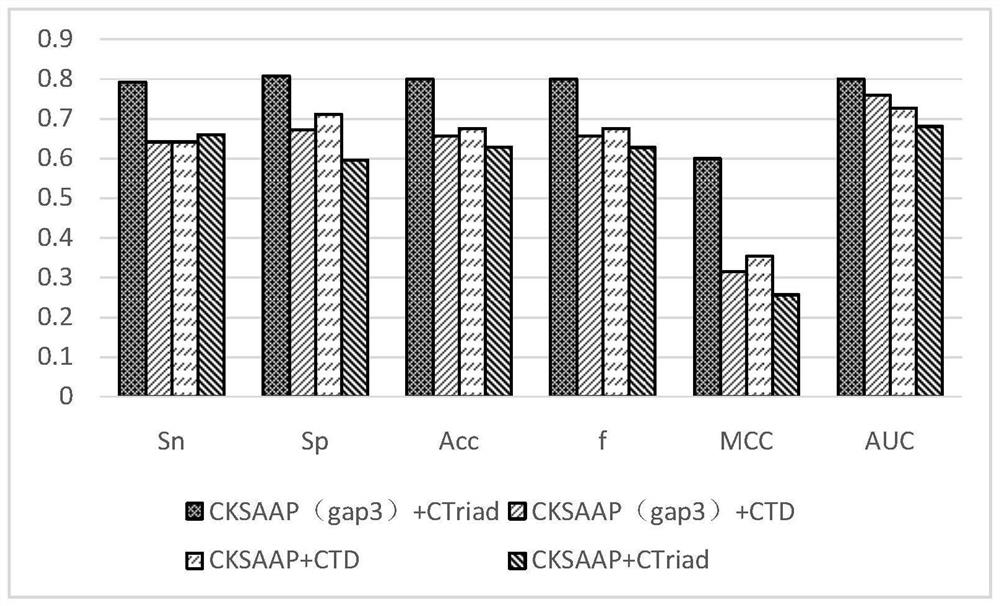
[0052] S2. Using CKSAAP and CT features to extract the vector features of the antioxidant protein data set to obtain two feature files, and splicing the two feature files to obtain a complete feature set.
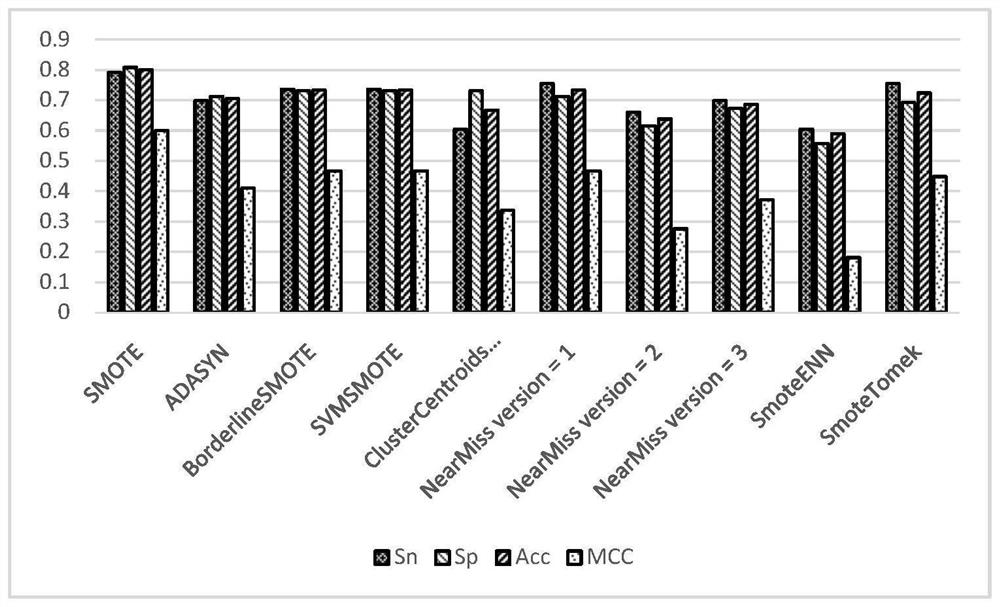
[0053] S3. Use a variety of unbalanced processing algorithms to perform unbalanced processing on the training set data, so that the data set reaches a balanced state;
[0054] S4. Using the MRMD algorithm to perform feature selection on the feature set to obtain fea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com