Sample pathogenic bacterium typing method and system
A pathogenic bacteria and sample technology, applied in the field of pathogen detection, can solve the problems of limiting system operating speed, slow operating speed, and limiting the application speed of cgMLST, and achieve the effect of improving system robustness and computing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] The following drawings, embodiments of the present application is explained in detail with reference to.
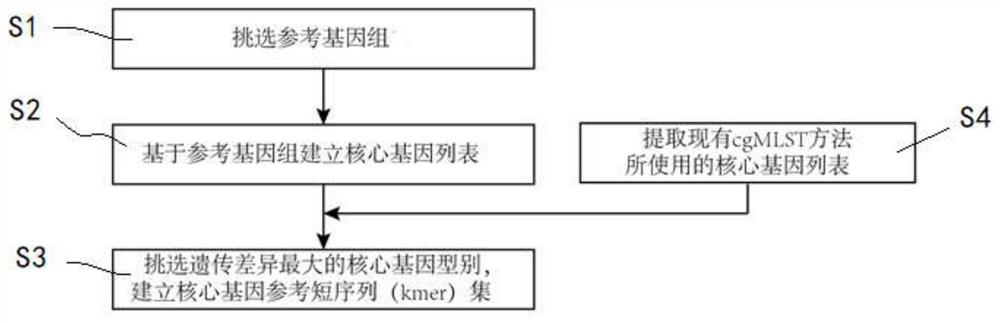
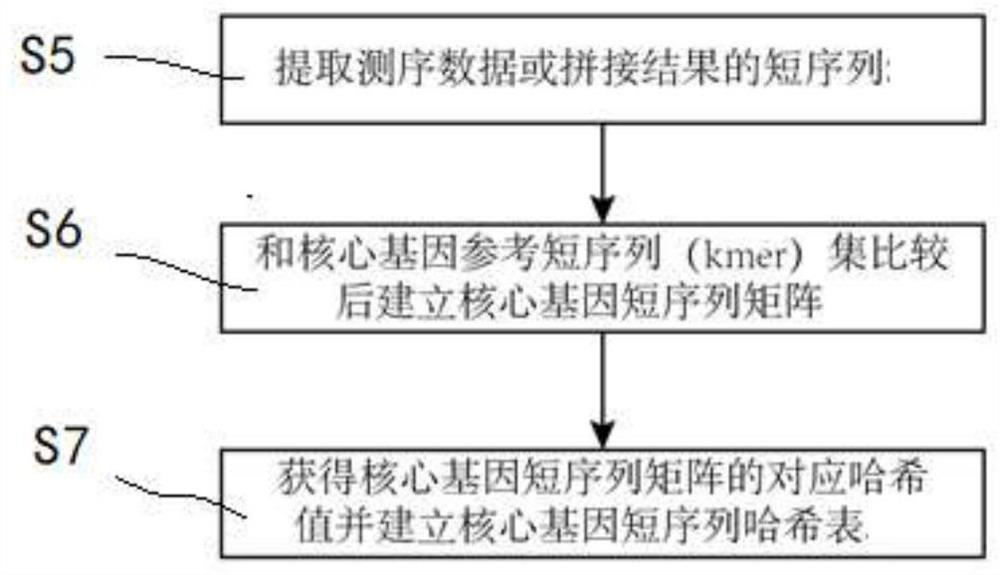
[0030] figure 1 Set is a flowchart illustrating example provided core gene construct of the invention with reference to a short sequence (kmer), the method may be performed by a processor in the form of a computer program. The method includes the following steps:
[0031]Establishing reference genome datasets step S1. To establish a genetic component type process for a certain pathogenic bacteria, first need to establish the kind of the reference genome datasets pathogenic bacteria. Reference genome data set consists of a series of the bacterial genome, of which genome data source can be downloaded directly from the common property genomic databases (e.g., GenBank, pubMLST, EnteroBase etc.), or from the sequencing results a database of public property (e.g., NCBI SRA, EBI ENA, etc.) after downloading the sequencing results obtained splicing, or get through splicing of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com