Construction method and device of plant metabolite database, medium and terminal
A construction method and metabolite technology, which can be used in database management systems, chemical informatics data warehouses, chemical information database systems, etc. Information-rich, easy-to-use effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
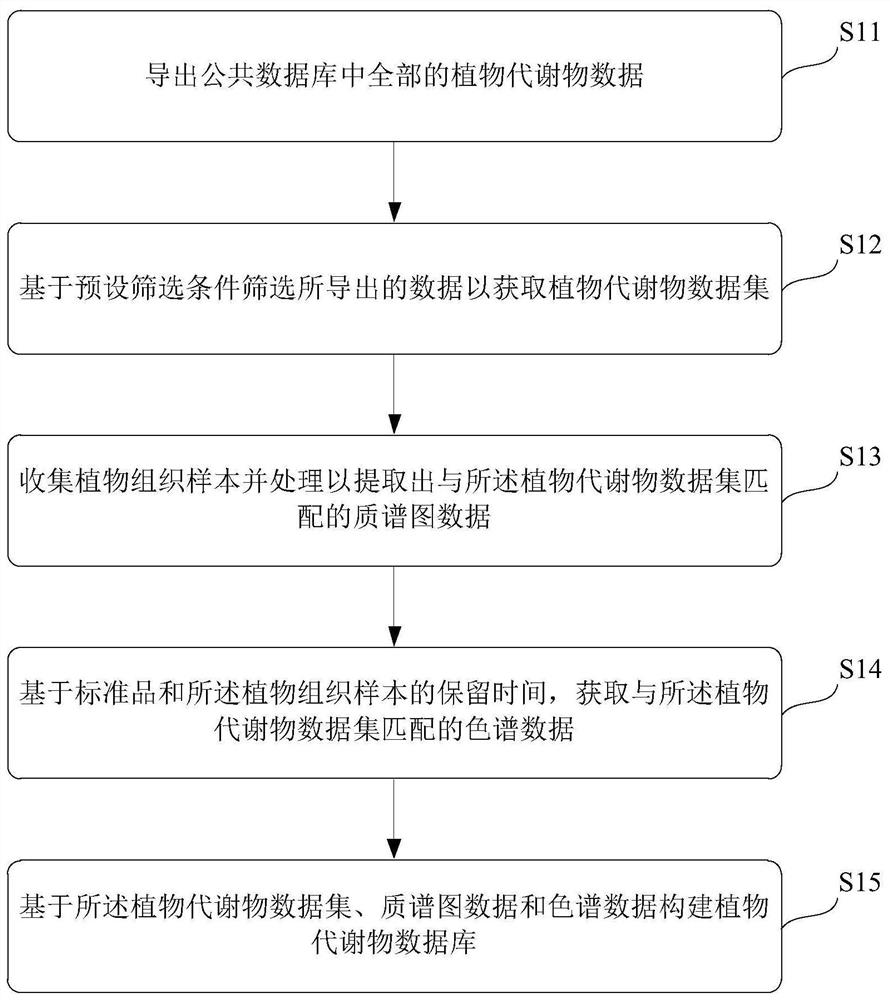
[0034] Such as figure 1 As shown, the embodiment of the present invention proposes a schematic flow chart of a method for constructing a plant metabolite database, which includes:
[0035] Step S11. Export all plant metabolite data in the public database. Optionally, the public database can be Metlin, HMDB, MassBank, etc. Take HMDB as an example, use Python to export the information of each level of all metabolites on the HMDB website; use Python to find the information of the Disposition layer corresponding to each metabolite, find the Biological column in this layer of information, and confirm whether the compound contains plant Information, if there is, all the information corresponding to the compound is exported, and the compound information meeting the above conditions is collected to form a list.
[0036] Step S12. Filter the exported data based on preset filter conditions to obtain a plant metabolite data set. Specifically, the derived data are screened based on the...
Embodiment 2
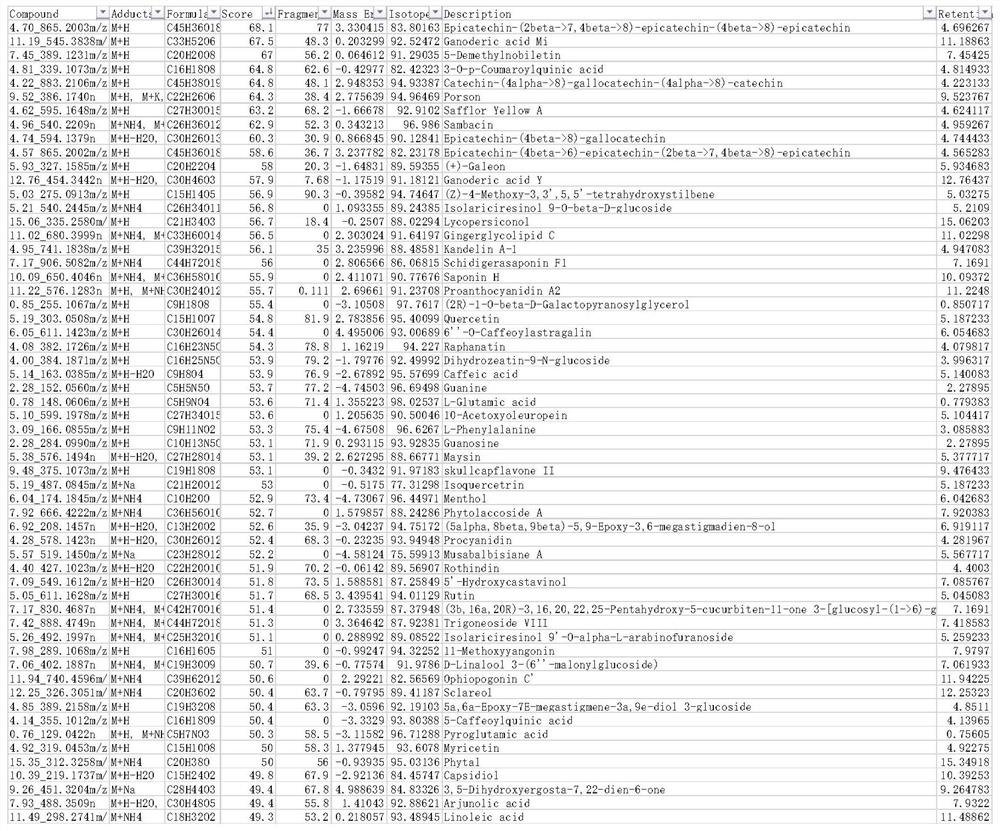
[0066] Such as Figure 6 As shown, the embodiment of the present invention provides a schematic structural diagram of a device for constructing a plant metabolite database. The device for constructing the plant metabolite database provided in this embodiment includes: an export module 61, which is used to export all plant metabolite data in the public database; a screening module 62, which is used to filter the derived data based on preset screening conditions to obtain plant Metabolite data set; Spectrogram data acquisition module 63, used to collect plant tissue samples and process to extract mass spectrum data matched with the plant metabolite data set; The retention time of the plant tissue sample is used to obtain chromatographic data matching the plant metabolite data set; the construction module 65 is used to construct a plant metabolite database based on the plant metabolite data set, mass spectrogram data and chromatographic data.
[0067] It should be noted that the...
Embodiment 3
[0070] The embodiment of the present invention provides a computer-readable storage medium, on which a computer program is stored, and when the computer program is executed by a processor, the method for constructing the plant metabolite database is implemented.
[0071] Those of ordinary skill in the art can understand that all or part of the steps for implementing the above method embodiments can be completed by hardware related to computer programs. The aforementioned computer program can be stored in a computer-readable storage medium. When the program is executed, it executes the steps including the above-mentioned method embodiments; and the aforementioned storage medium includes: ROM, RAM, magnetic disk or optical disk and other various media that can store program codes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com