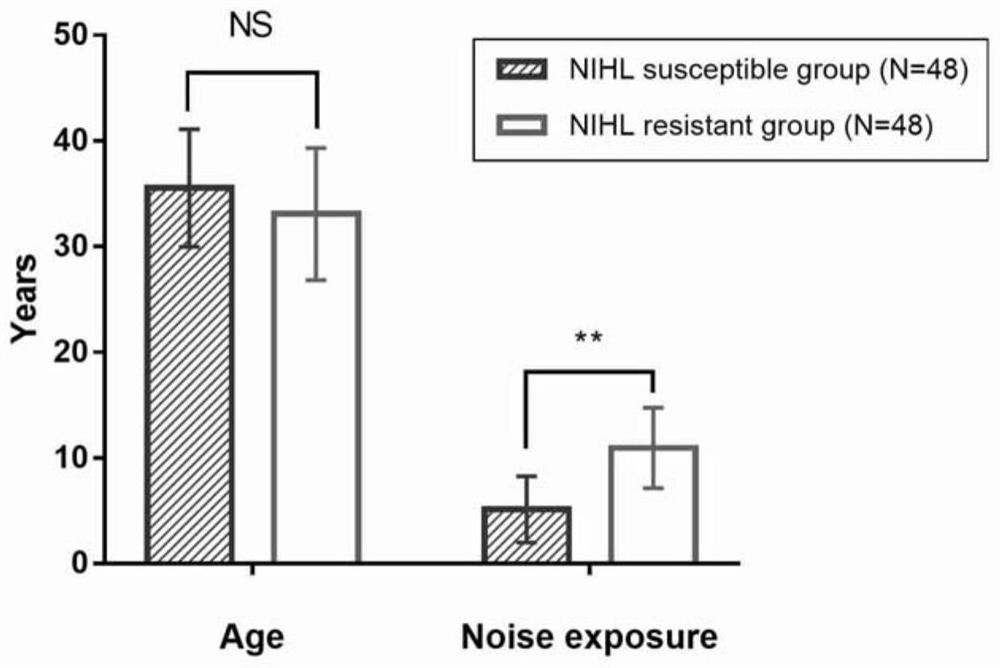
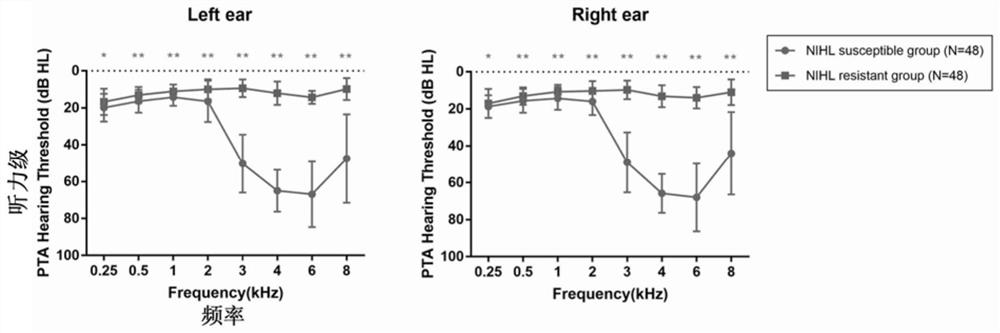
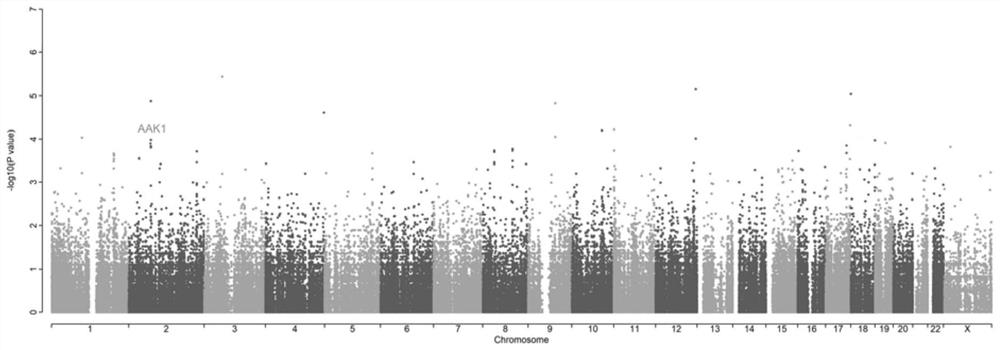
Biomarker for predicting susceptibility risk of noise-induced hearing loss and application thereof
A biomarker and hearing loss technology, applied in the field of biomarkers, can solve the problem that no research has clearly identified NIHL susceptibility-related gene loci
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] A method of predicting the risk of susceptibility to noise-induced hearing loss comprising the steps of:
[0051] (1) Extract DNA from the subject's whole blood sample;
[0052] (2) PCR instrument was used to amplify the rs1396793 polymorphic site in the 5'UTR region of gene AAK1, wherein the primer sequence was: 5'-GAATAGGGAGCAGCAAAGC-3'(forward), 5'-GAGCCGTCTCAAGTGGGT-3'(reverse) ;Reaction conditions: 98°C for 2min; 30cycles: 98°C for 10s, 58°C for 15s, 72°C for 30s; 72°C for 2min.
[0053] (3) sequencing the amplification primers;
[0054] (4) Analyze the genotype of the rs1396793 polymorphic site in the sequencing results. If it is AA, it indicates a homozygous mutation, and it is judged that the subject is a noise-induced hearing loss tolerant population; if the genotype is wild homozygous CC or heterozygous CA, it is judged that the subjects are susceptible to noise-induced hearing loss.
Embodiment 2
[0056] A kit for predicting the risk of susceptibility to noise-induced hearing loss, characterized by comprising: primers for amplifying the rs1396793 polymorphic site in the 5'UTR region of gene AAK1, wherein the primer sequence is: 5'- GAATAGGGAGCAGCAAAGC-3' (forward), 5'-GAGCCGTCTCAAGTGGGT-3' (reverse). The kit also contains other commonly used reagents for PCR amplification, such as dNTPs, polymerase, buffer, etc.
[0057] In summary, the biomarker rs1396793 provided by the present invention can be used to predict the risk of susceptibility to noise-induced hearing loss.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com