293A cell strain for producing adenovirus as well as preparation and application thereof
An adenovirus and cell technology, applied in the field of cell engineering, can solve the problems of RCA and HDEP pollution, achieve the effect of preventing RCA and HDEP and improving safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Obtaining of exogenous fragments
[0040]Gene fragments LoxM2 (nucleotide sequence shown in SEQ ID NO.1), HPRT intron (nucleotide sequence shown in SEQ ID NO.2) and hygro cassette (nucleotide sequence shown in SEQ ID NO.3) were synthesized respectively shown), the three gene fragments were ligated sequentially to obtain the exogenous fragment (the nucleotide sequence is shown in SEQ ID NO.5), and the gRNA recognition sequence (the nucleotide sequence is shown in SEQ ID NO. NO.4), to obtain an exogenous fragment containing the gRNA recognition sequence.
Embodiment 2
[0041] Example 2 Construction of recombinant plasmids
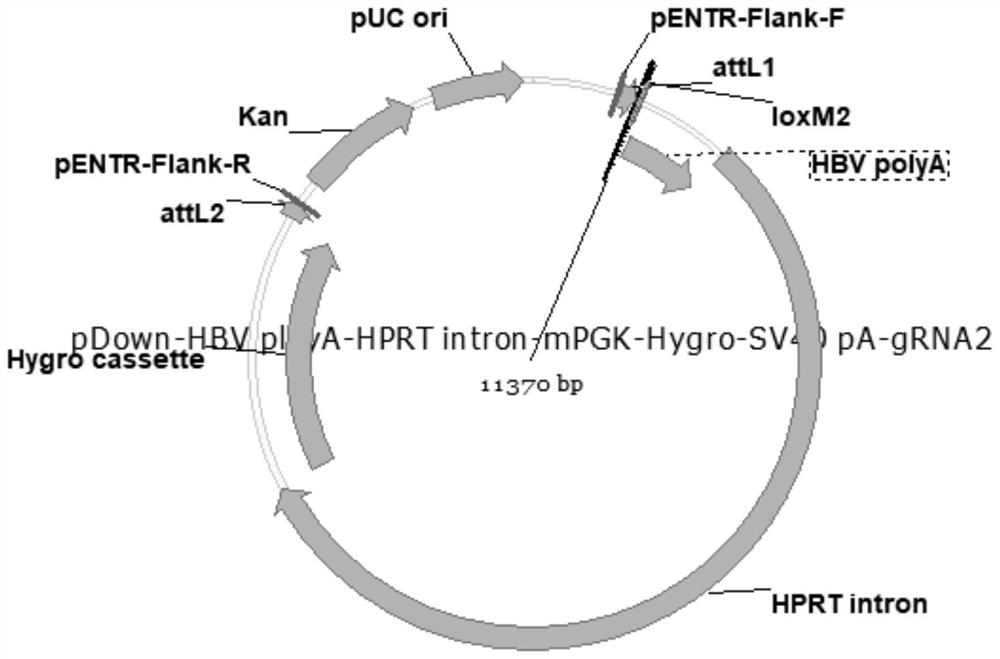
[0042] (1) Construct plasmid pDown-HBV ployA-HPRT intron-mPGK-Hygro-SV40 pA-gRNA2
[0043] The foreign fragment containing the gRNA recognition sequence obtained in Example 1 was cloned into the plasmid pDown-SapI-ccdB-Chl-SapI to obtain the recombinant plasmid pDown-HBV ployA-HPRT intron-mPGK-Hygro-SV40 pA-gRNA2, which Recombinant plasmid map such as figure 1 shown.
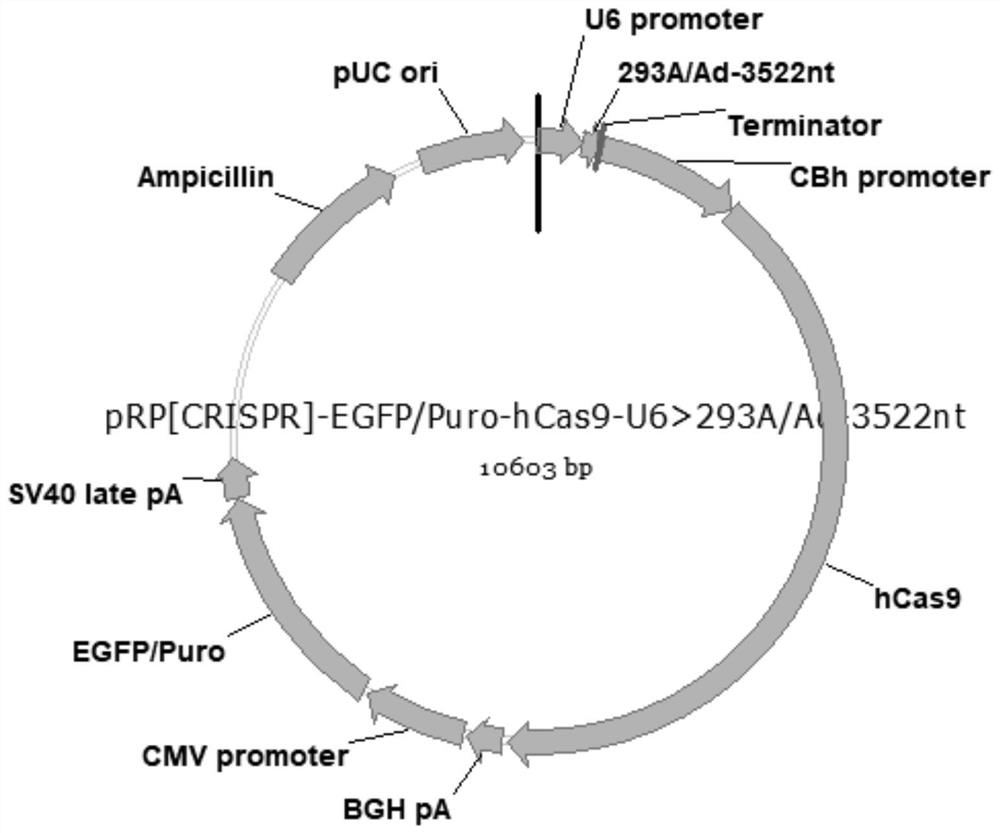
[0044] (2) Construction of plasmid carrying hCas9 and gRNA
[0045] Synthetic gene fragment hCas9 (nucleotide sequence shown in SEQ ID NO.6) and gRNA transcription frame (nucleotide sequence shown in SEQ ID NO.7), gRNA sequence is cloned into plasmid PX330-U6-BbsI-CBh -hSpCas9-CMV>EGFP / Puro obtained the recombinant plasmid pRP[CRISPR]-EGFP / Puro-hCas9-U6>{293A / Ad-3522nt}, the recombinant plasmid map is as follows figure 2 shown.
Embodiment 3
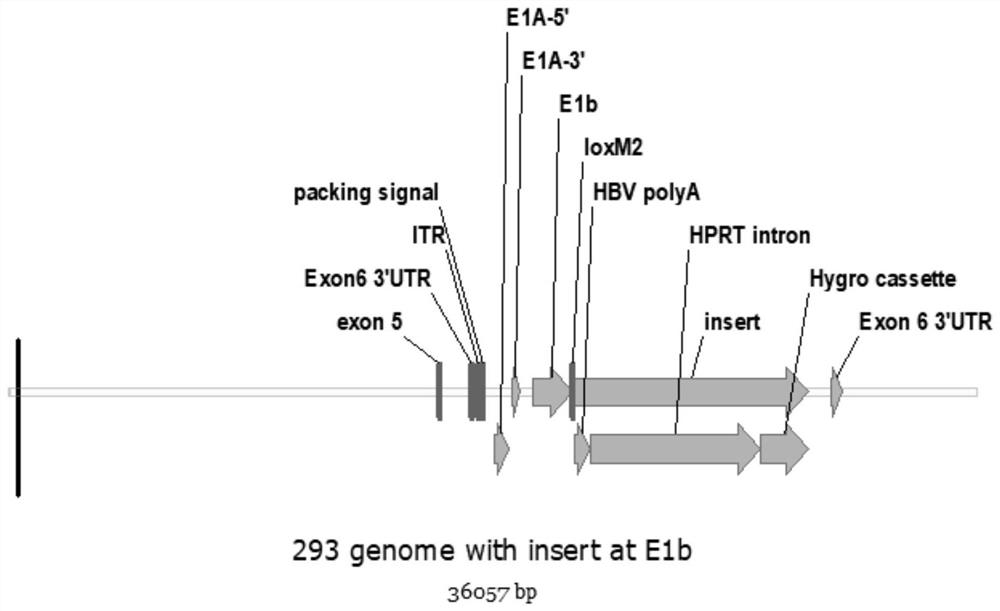
[0046] Example 3 Construction of VB-293A cell strain
[0047] Use 3×10 6 Cells were plated in a 6-well plate, and the confluence of the cells reached 80%-90% the next day. The two plasmids constructed in Example 2 were co-transfected into 293A cells (Thermo Fisher product number: R70507) with Lipofect amine3000 transfection reagent. According to the principle of NHEJ, hCas9 / gRNA will simultaneously recognize and cut the target site of the gene and the two target sites of the plasmid pDown-HBV ployA-HPRT intron-mPGK-Hygro-SV40 pA-gRNA2. The linearized exogenous fragment is inserted into the targeted site in the genome of the cell, resulting in targeted integration. The total amount of transfected plasmids is 2.5μg, plasmid pRP[CRISPR]-EGFP / Puro-hCas9-U6>{293A / Ad-3522nt}: 1.7μg; plasmid pDown-HBV ployA-HPRT intron-mPGK-Hygro-SV40 pA - gRNA2: 0.8 μg; Lipofectamine 3000: 7.5 μL. Change the solution every other day.
[0048] Passage the cells 48 hours after transfection to a 6...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com