Full-automatic quantitative analysis method for perinuclear lysosome distribution
A quantitative analysis, lysosome technology, applied in the field of cell analysis, can solve problems such as difficult to achieve high-throughput big data analysis, and achieve the effect of computational robustness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
no. 1 example
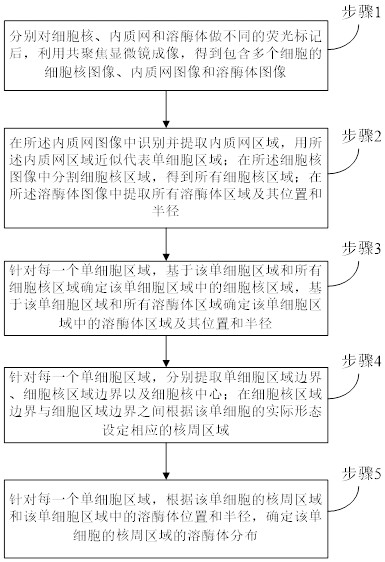
[0040] In the first embodiment of the present invention, a nuclear cycosocidizer distribution automatic quantitative analysis method, such as figure 1 As shown, the following specific steps are included:
[0041] Step 1, after different fluorescence markers of the nucleus, the nucleus microscopy is used to obtain a nucleus image, an endometric web image, and a lysosomas image including a plurality of cells.
[0042] Step 2, identify and extract the endoplasmic network area in the inner textone web image, and use the inner texture network area to represent a single cell region; in the nuclear image, the nucleus is divided into all nuclear region; All lysosomas are extracted in the lysosomal image and its position and the radius thereof;
[0043]Alternatively, in the step 2, a depth learning-based instance division model MASKRCNN is identified and extracted in the endoplasmic web region, and the endogenic network region is used to approximate a single cell region;
[0044] In the st...
no. 2 example
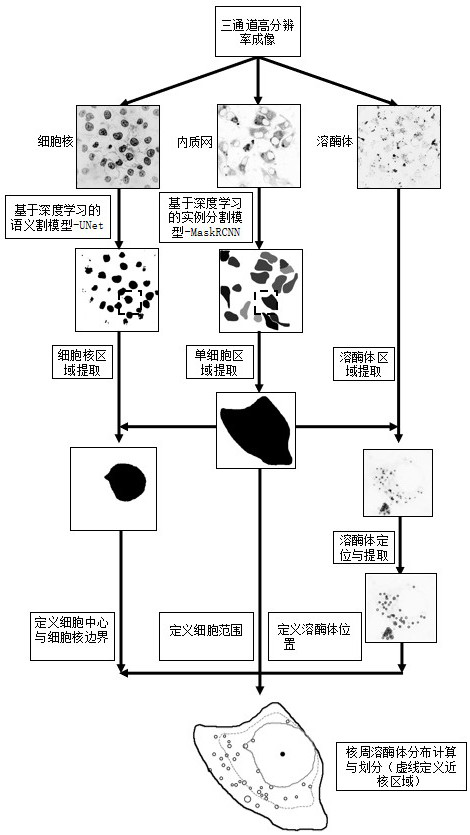
[0066] In the second embodiment of the present invention, the present embodiment is incorporated herein by reference. Figure 2 ~ 3 An application example of the present invention is described.
[0067] like image 3 As shown, the nuclear cycosome distribution automatic quantitative analysis method according to the embodiment of the present invention, including the following specific steps:
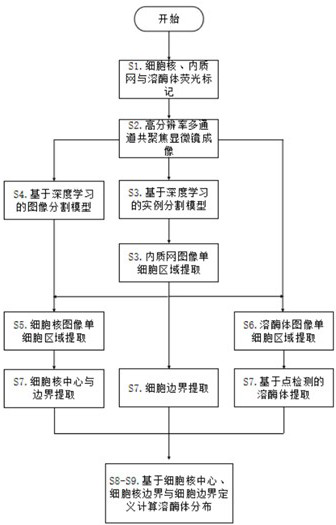
[0068] Step S1: Different fluorescent markers are made to the nucleus, endometric webs and lysosomes.
[0069] Step S2: Through high-resolution multi-channel copolymerization microscopy, it is obtained from nuclear, endometric webs and lysosomas, each image containing multiple cells, generally more than 10, such as figure 2 Indicated.
[0070] Step S3: By using the inner texture network, the characteristics of the entire cell region are occupied by the internal quality web image. Use the depth learning-based instance division model Maskrcnn to identify and extract the endometric network area, a...
no. 3 example
[0081] According to a third embodiment of the present invention, the flow of nuclear cyclic exclusive distribution automatic quantitative analysis method is the same, and the second embodiment is the same, and the present embodiment can be used in the project implementation. The general purpose hardware platform is implemented, of course, can also pass hardware, but in many cases, the former is better implementation. Based on this, the method of the present invention can be embodied in the form of a computer software product, which is stored in a storage medium (such as ROM / RAM, disk, disc), including several instructions to make one The method described in the embodiment of the present invention is performed.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com