Parameter-free nonlinear intelligent optimization method for identifying cancer driving pathway
An intelligent optimization and nonlinear technology, applied in the field of bioinformatics, can solve the problems of cumbersome, weak scalability, etc., to achieve the effect of more useful information, fast solution speed, high recognition efficiency and practical value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] A non-parametric nonlinear intelligent optimization method for identifying cancer-driving pathways, comprising the following steps:
[0066] 1) Set a nonlinear model without artificial parameters:
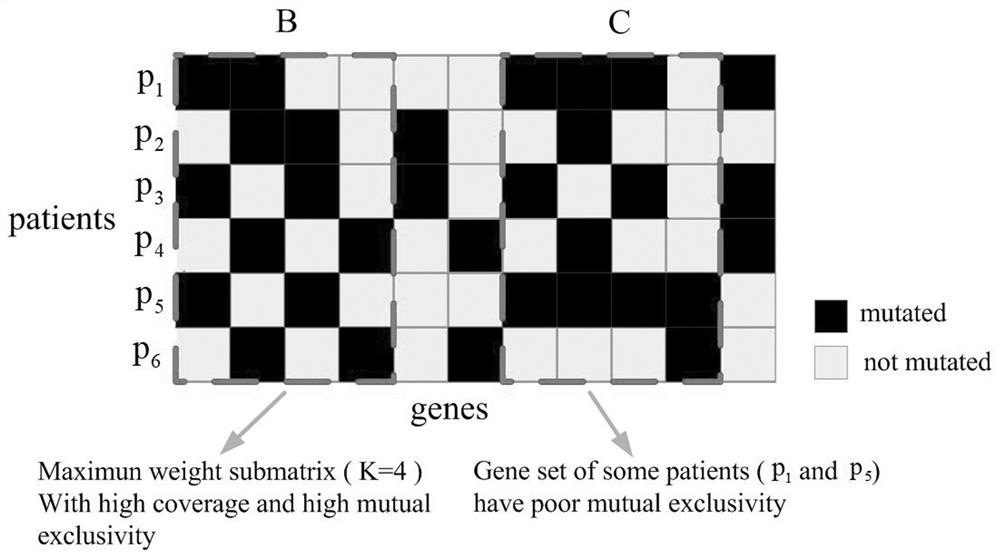
[0067] Using the existing somatic mutation matrix, copy number variation matrix and protein interaction network, where the somatic mutation matrix is denoted as The copy number variation matrix is denoted as The rows of matrix S and matrix C represent the same sample set P, and the columns represent the gene set G respectively S and G C , the value of each element in matrix S is S∈{0,1}, if gene j is mutated in patient i, then S ij = 1, otherwise S ij =0, the values of the elements in the matrix C are C∈{-2,-1,0,1,2}, where the values of the elements in the copy number variation matrix C are obtained by analyzing the GISTIC tool, set Q=(V,E) Represents a connected PPI network where each vertex v i ∈V denotes the gene g i expressed protein, each undirected ed...
Embodiment 2
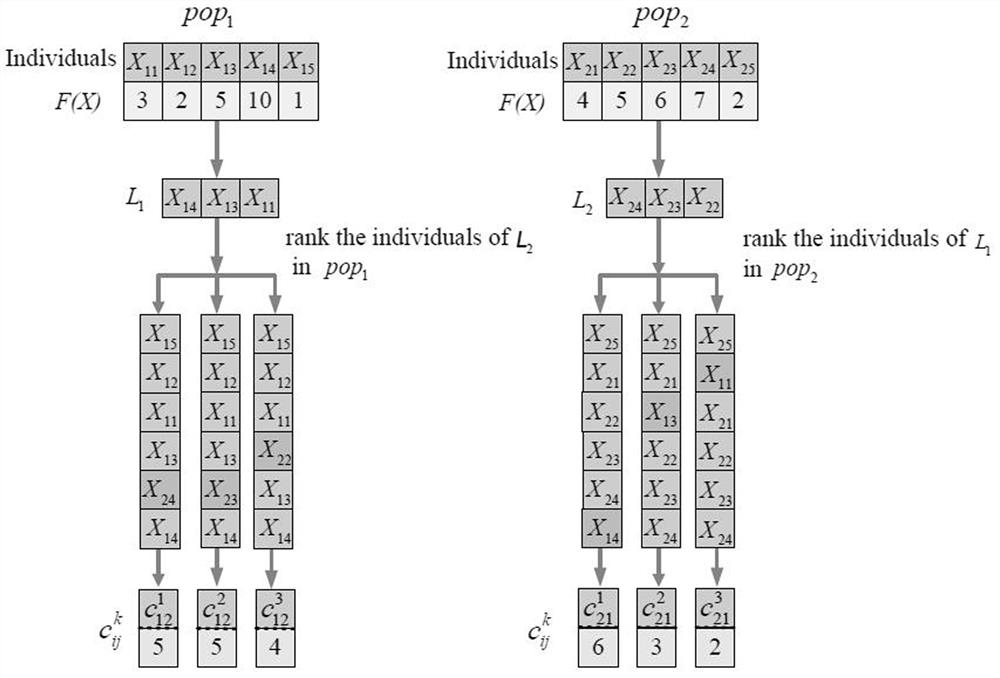
[0111] Step 6) input mutation matrix A p×G , where |P|=90, |G|=440, PPI network Q, there are 9859 vertices and 40705 edges in the network, set the driving path size K=7, and then input CCA-NMWS related parameters: population size N= log2(|G|^K)*2=122, mutation probability P m1 = 0.5 and P m2 =0.8, maximum evolution generation maxg=1000, threshold maxt=10 to keep the optimal value constant; every subpopulation evolves T=10 generations to judge whether to compete, and the proportion of opponent set=0.2;
[0112] Step 8) Obtaining the size of the drive path of K=7, the rest of the steps are the same as in Embodiment 1, such as Figure 4 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com