Method for detecting tumor cells or tumor cell debris
A tumor cell and debris technology, applied in the field of detecting tumor cells or tumor cell debris by enhancing the detection signal-to-noise ratio, and detecting tumor cells or tumor cell debris, can solve the problems of not excluding white blood cells, weak CTC detection signal, and low efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0121] Example 1: Comparison of the expression levels of ALPL, ALPP, ALPI and ALPG genes in tumor cells and white blood cells (WBC)
[0122] 1.1 Experimental steps
[0123] (1) Extract total RNA from white blood cells (WBC) and tumor cell lines Lovo, MCF-7, SW480, HCC827, H820, MKN 7, 7721, Caco2, and Bewo (Table 1), and degDNA and reverse the RNA samples recording process to obtain cDNA.
[0124] Table 1 Cell line names and their corresponding tumor types
[0125]
[0126] (2) The cDNA obtained in step (1) was quantitatively amplified by qPCR according to the following system (Table 2 and 3), to detect WBC and tumor cells Lovo, MCF-7, SW480, HCC827, H820, MKN 7, 7721, The average CT value of Caco2, and Bewo, and the obtained average CT value was calculated by 2–ΔΔCT (Livak) method to obtain the relative value of WBC and tumor cell lines MCF-7, SW480, HCC827, H820, MKN 7, 7721, Caco2, and Bewo The relative expression of ALPL, ALPP gene, ALPI gene and ALPG gene in Lovo ce...
Embodiment 2
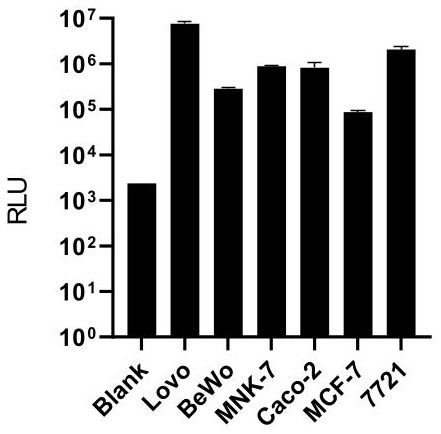
[0142] Example 2: Activity Test of Tumor Cell Line Alkaline Phosphatase
[0143] 2.1 Experimental steps
[0144] (1) Cell preparation: Digest the tumor cell lines Lovo, Bewo, MKN-7, Caco-2, MCF-7, and 7721 with 0.25% trypsin for 2 minutes, then stop the digestion with 1640 complete medium, and then use 1× Wash the cells with PBS, centrifuge to remove the supernatant, resuspend the cells with 1ml 1×PBS and count with a hemocytometer. 1.5ml EP tubes containing 1000 Lovo cells (Lovo-1000), 1.5ml EP tubes containing 1000 BeWo cells per tube (BeWo-1000), 1000 MKN-7 cells per tube (MKN-7-1000 ), 1.5ml EP tubes containing 1000 Caco-2 cells per tube (Caco-2-1000), 1.5ml EP tubes containing 1000 MCF-7 cells per tube (MCF-7-1000) , 1.5ml EP tube containing 1000 7721 cells (7721-1000) per tube.
[0145] (2) Chemiluminescence detection:
[0146] test group:
[0147] Add alkaline phosphatase chemiluminescence substrate solution (alkaline phosphatase substrate solution A04, purchased f...
Embodiment 3
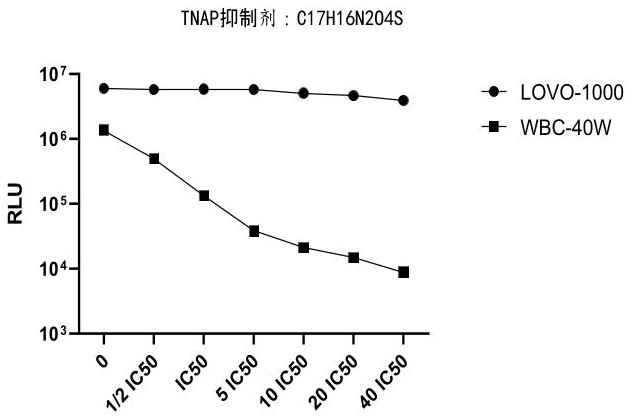
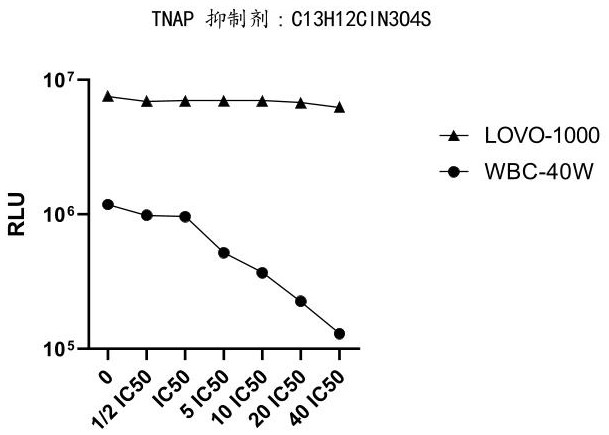
[0152] Example 3: Effect of TNAP inhibitors on increasing the ratio of tumor cell lines to leukocyte chemiluminescence readouts
[0153] In this embodiment, blood samples are used as samples to be tested for related experiments.
[0154] 3.1 Experimental steps
[0155] (1) WBC cell preparation:
[0156] Take 2ml of blood, lyse with 20ml of 1×red blood cell lysate for 10min, centrifuge at 500g for 5min, remove the supernatant, then wash twice with 1×PBS, centrifuge at 500g for 5min after the first wash, remove the supernatant, and then wash for the second time , Centrifuge again at 300g for 3min, remove the supernatant, resuspend the cells with 1ml 0.5% BSA, obtain WBC cell suspension, and count with a hemocytometer. Add the cell suspension to the 1.5ml EP tube according to the amount of 400,000 white blood cells (WBC-40W) per tube to obtain 1.5ml EP tubes containing 400,000 white blood cells (WBC-40W) per tube.
[0157] (2) Lovo cell preparation:
[0158] Digest Lovo cells w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com