Method and device for developing disease marker by using immunoglobulin-associated proteome
An immunoglobulin and proteome technology, applied in the field of developing disease markers, can solve the problems of few antigen identities and elusive immunoglobulin homology characteristics, and achieve the effect of reducing complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1, Quantitative analysis of immunoglobulin-associated proteome (IgAP)
[0067] Serum samples from 38 AMI patients, 20 stable CAD patients, and 37 individuals diagnosed with non-CAD / AMI (NCA) disease constituted a discovery cohort. Table 1 summarizes the clinical characteristics of patients with CAD and AMI.
[0068] Table 1. Demographic characteristics of the MS dataset
[0069]
[0070]
[0071] *± indicates standard error (SE). / indicates a data gap.
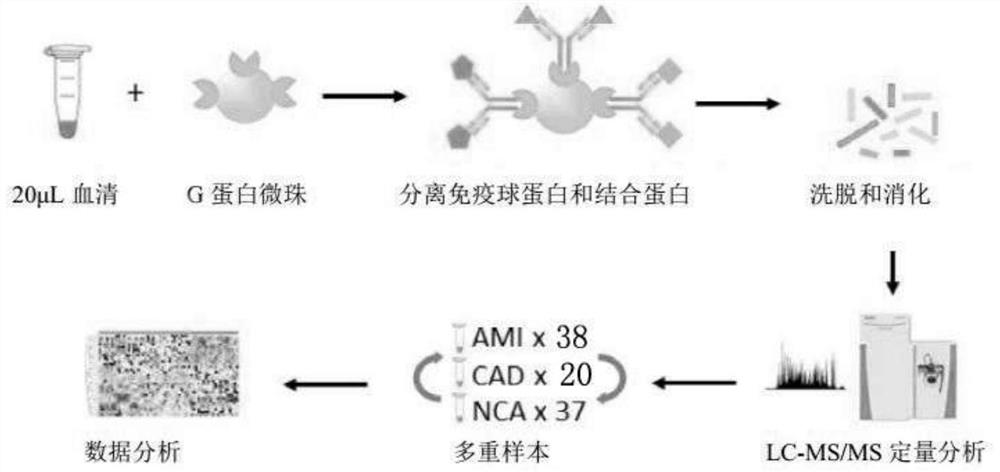
[0072] To study the immunoglobulin-associated proteome (IgAP), this example employs an IgAP assay that integrates purification of immunoglobulin complexes with label-free quantitative LC-MS / MS, as figure 1 As shown, the method includes:
[0073] According to the operation described in the aforementioned "1.3 IgAP isolation", the immunoglobulin-associated proteome with protein G agarose beads was isolated and purified from 20 μL serum of each individual using Protein G magnetic beads;
[0074] Accordi...
Embodiment 2
[0080] Embodiment 2, distinguish the CAD of AMI patient by IgAP
[0081] The present invention assessed whether IgAP profiles could differentiate AMI, CAD and NCA individuals using unsupervised hierarchical cluster analysis. As a result, 95 patient samples were classified into four main classes, which were named classes I to IV ( Figure 5 ). CAD individuals clustered well in group IV and were completely separated from AMI cases, which were distributed in groups II (21 cases) and III (16 cases), respectively. The complete separation of the CAD group from the AMI patients indicated a large difference between the IgAP profiles of CAD and AMI patients. As for the NCA samples, while cluster I contained only 18 NCA samples, clusters II, III, and IV contained 10, 5, and 4 NCA samples, respectively ( Figure 5 ). NCA patients came to the clinic for different reasons, so their unique clustering may be due to indeterminate clinical features independent of CAD / AMI.
Embodiment 3
[0082] Example 3, Differential enrichment of IgAP protein in patient population
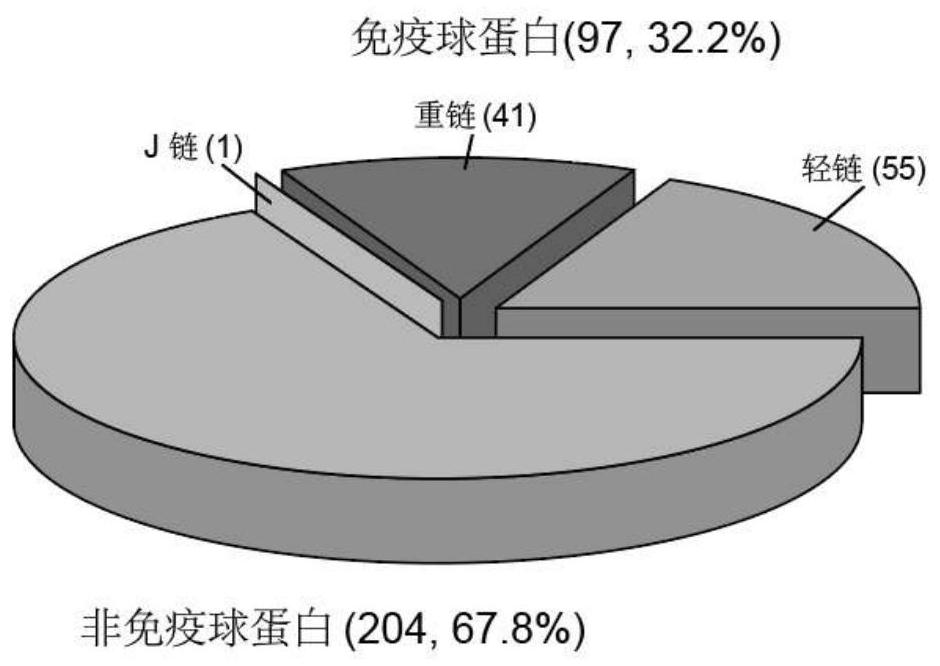
[0083] Differential enrichment of IgAP protein between patient groups Complete isolation of CAD from AMI samples suggests that the presence of IgAP protein can differentiate atherosclerotic patients with and without myocardial infarction. Therefore, the present invention performed a pairwise comparison between IgAP proteins. Three patient groups. Comparing all AMI patients with CAD patients, 17 IgAP proteins were found to be significantly elevated in AMI patients ( Figure 6A-6B ). Gene Ontology (GO) analysis of these AMI-elevated IgAP identified complement and coagulation cascade pathways (FDR = 1.24 × 10 -6 ), where different complements (C1s and C1r) and coagulation factors (F10 and F5) represent proteins ( Figure 6A-6B ). Several variable regions of immunoglobulins (IGKV6D-21, IGHV1-69, IGKV1-12, IGHV1-46) were also elevated in IgAP of AMI patients. This result suggests that immunoglob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com