Application of miRNA molecular marker in schizophrenia detection
A molecular marker, schizophrenia technology, applied in the direction of determination/inspection of microorganisms, biochemical equipment and methods, etc., to achieve the effect of highlighting substantive characteristics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Acquisition and functional annotation of differentially expressed genes;
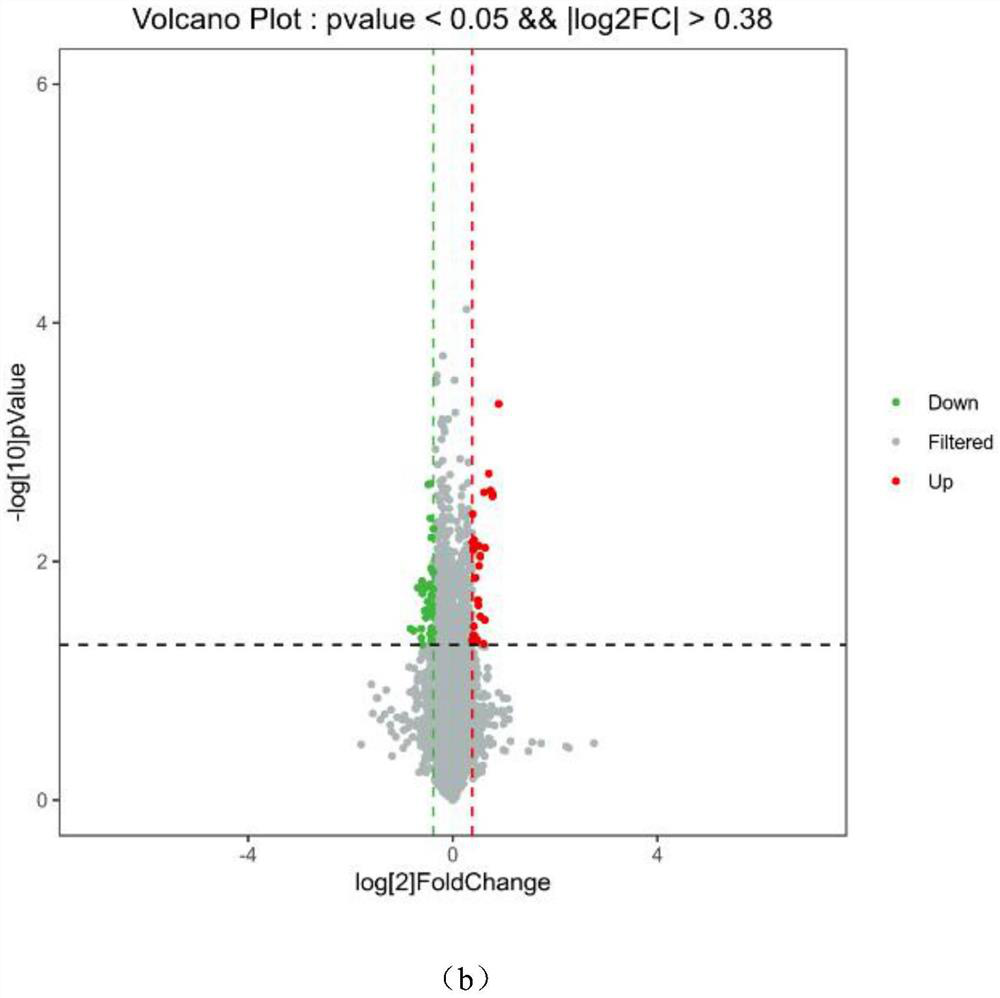
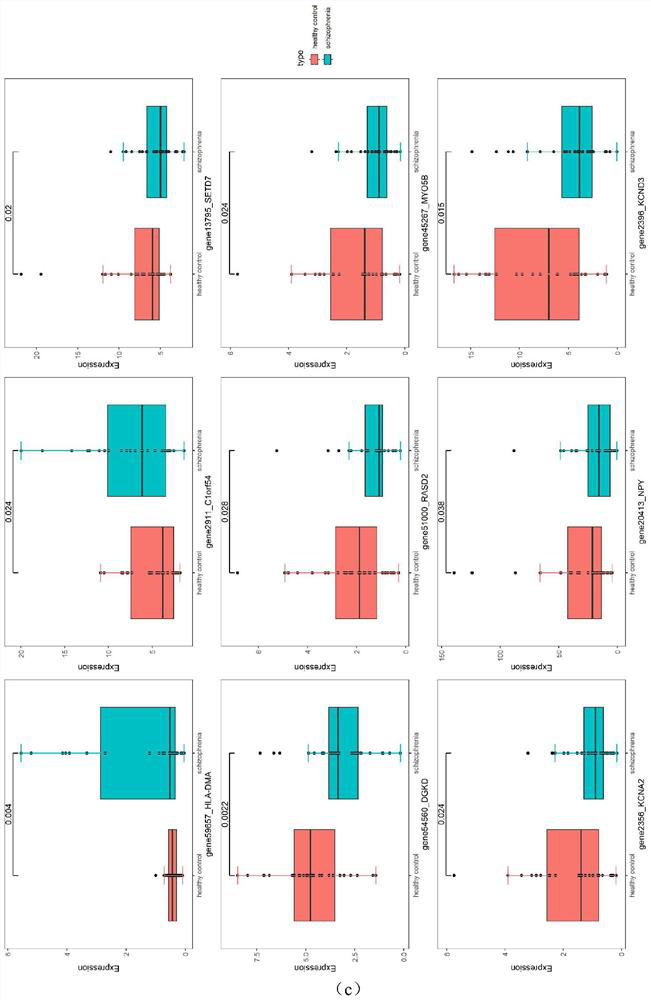
[0041] To identify the differentially expressed genes between schizophrenia patients and healthy individuals, the previously reported RNA-Seq data (GEO accession number GSE121376) was obtained. Use the locally developed Perl script to delete the low-quality bases at the 5' and 3' ends (Q20 ≤20). The read fragment was aligned to the human reference genome (GRCm38.p5) using HISAT 2 (version 2.0.5). BEDTools was used for reading and counting (Quinlan 2014), and RPKM (Reading Per Million Bases) method was used to calculate its expression level. The differentially expressed genes were screened by using the multiple change threshold of |log2FC|>0.4, and the P value was obtained by T-test. Subsequently, DAVID 6.8 was used for functional enrichment analysis of schizophrenia-related genes.
Embodiment 2
[0042] Example 2: Construction of ceRNA network:
[0043] Collect miRNA from miRbase. The mRNA and lncRNA sequences were obtained from NCBI(GRCm38.p5). All differentially expressed mRNA and lncRNA were used for target identification. The regulatory relationship between miRNA-mRNA and miRNA-lncRNA was determined by RNA in situ hybridization (set the coordinates of seed region to 1 to 6, 2 to 7, 3 to 8, 4 to 9 and 5 to 10; Mfe≤-25kcal / mol, p≤0.01) and BLASTN(E value is 1000, except miRNA and miRNA in forward alignment (alignment position is greater than 6), and the intersection point is obtained. Use Cytoscape(3.5.1) to visualize ceRNA network.
Embodiment 3
[0044] Example 3: Establishment of Schizophrenia Mouse Model:
[0045] The N- methyl -D- aspartic acid (NMDA) glutamate receptor antagonist MK-801(Sigma-Aldrich, St.Louis, MO., USA) was dissolved in physiological saline and injected into mice by intraperitoneal injection for 7 days (0.6mg / kg once a day). Mice under the same conditions were injected with the same dose of normal saline as control. PET and qPCR experiments were performed within 1 week after injection.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com