Application of SNP (Single Nucleotide Polymorphism) marker in inbred line mouse strain identification and primer sequence
A primer sequence and inbred line technology, applied in the direction of recombinant DNA technology, biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of identification of inbred line mice, differences in epigenetic traits, etc., to achieve The effect of saving time spent on identification, simple operation, and improving detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example 1-15
[0136] Preparation Examples 1-15 respectively provide a pair of primer sequences. The difference between each preparation example is that the SNP sites used for amplification are different. The details are shown in Table 1.
[0137] The design method of primer sequence specifically comprises the following steps:
[0138](1) Find the upstream and downstream nucleotide sequences of the SNP site on the Ensembl website (http: / / asia.ensembl.org / index.html), and use the online primer design website Primer blast (https: / / www.ncbi.nlm. nih.gov / tools / primer-blast / index.cgi LINK_LOC=BlastHome) to design the primer sequences for PCR amplification of each SNP locus. The design results of the primer sequences are shown in Table 1. Among them, the rs (reference snp) number is a certain mutation that occurs at a certain point of the gene, and the mutation sites of these characteristics are encoded in the Human Genome Project, which is the rs number.
[0139] (2) After the design of the pr...
Embodiment 1
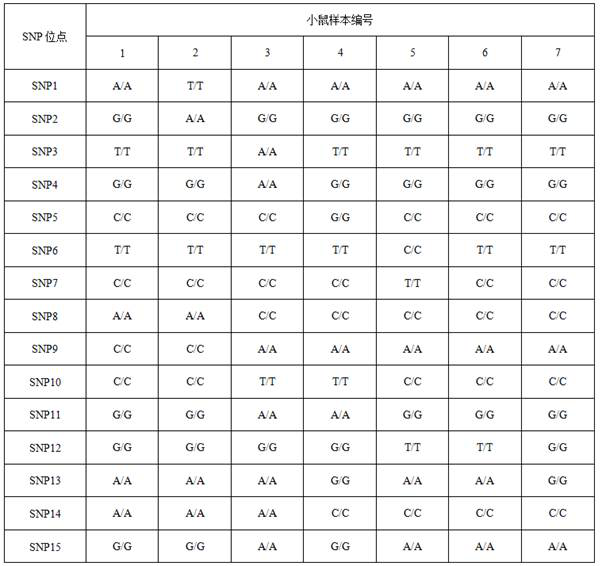
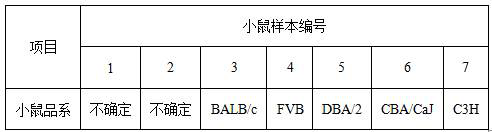
[0144] This example provides a method for identifying inbred mouse strains. There are 7 kinds of mouse samples identified in this embodiment, corresponding to different mouse strains. The above mouse samples were all obtained from SPEF (Beijing) Biotechnology Co., Ltd. The types of 15 SNP loci known in 7 mouse strains are shown in Table 2.
[0145] Table 2 Types of 15 SNP sites in 7 mouse strains
[0146]
[0147] The identification method of inbred mouse strain specifically comprises the following steps:
[0148] 1. Use a genomic DNA extraction kit to extract genomic DNA. The genomic DNA extraction kit used is a blood / cell / tissue genomic DNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., DP304). Specifically include the following steps:
[0149] (1) Put the 2mm mouse tail tip into a 1.5mL sterile centrifuge tube, add 200μL Buffer GA and 10μL proteinase K, place it in a water bath at 56℃ for 12h at constant temperature, and lyse;
[0150] (2) Conti...
Embodiment 2
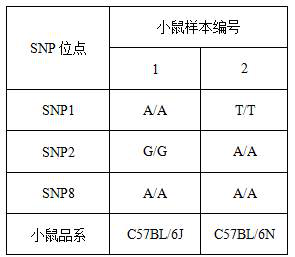
[0172] This example provides a method for identifying inbred mouse strains. The difference between this example and Example 1 is that there are 2 mouse samples identified in this example. It is known that the mouse samples are two mouse strains, C57BL / 6J and C57BL / 6N, and it is necessary to identify the mouse samples Specific corresponding mouse strains. The above mouse samples were all obtained from SPEF (Beijing) Biotechnology Co., Ltd. Other operations are all the same as in Example 1.
[0173] According to the comparison of SNP loci between the two mouse strains C57BL / 6J and C57BL / 6N in Table 2, it can be seen that the types of SNP1, SNP2 and SNP8 can be identified, and the mouse strain of the mouse sample can be determined. Therefore, only the PCR amplification of the above three SNP sites was performed.
[0174] The detection results and identification results of the 3 SNP sites of the 2 mouse samples in this example are shown in Table 5.
[0175] Table 5 The detecti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com