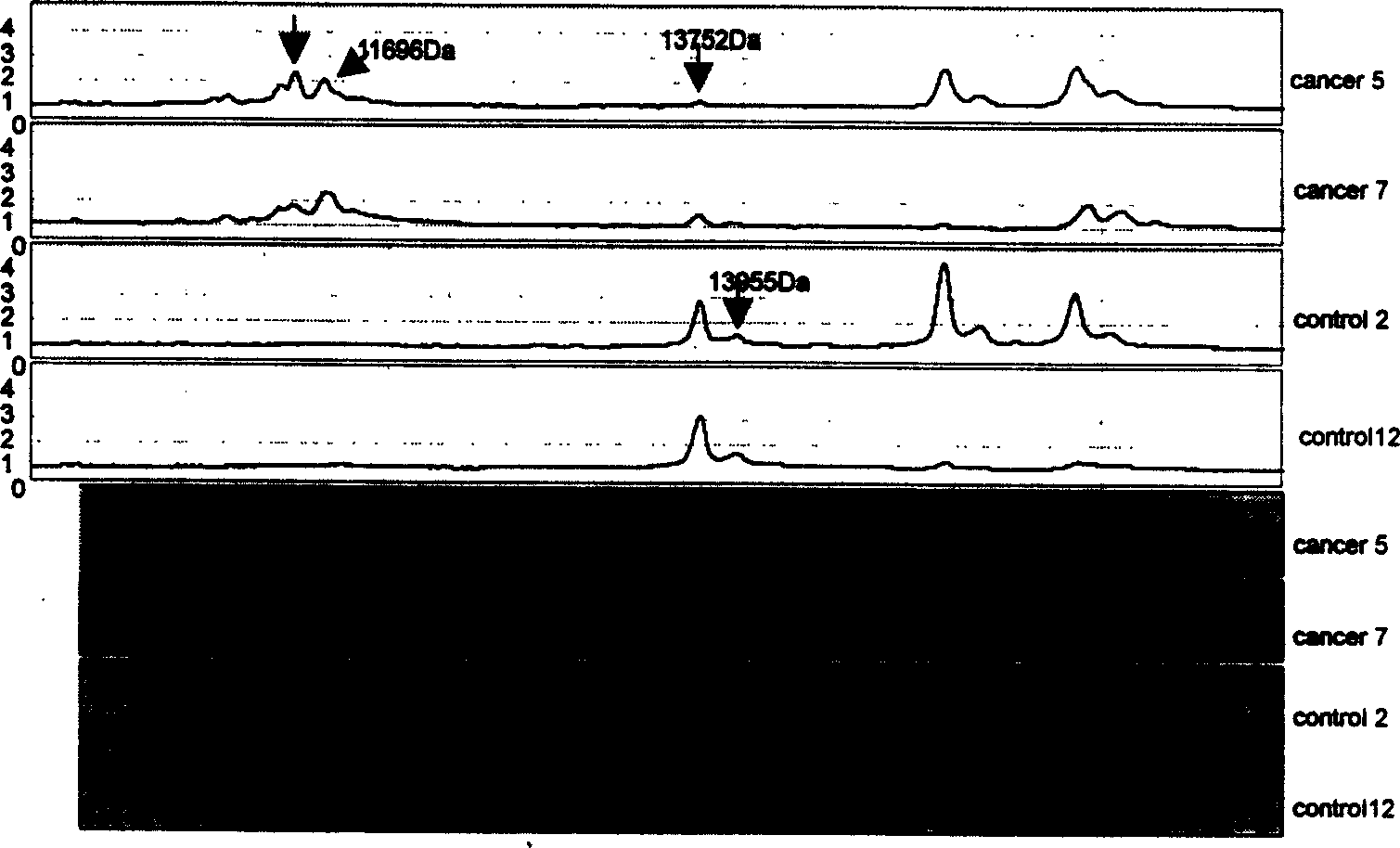
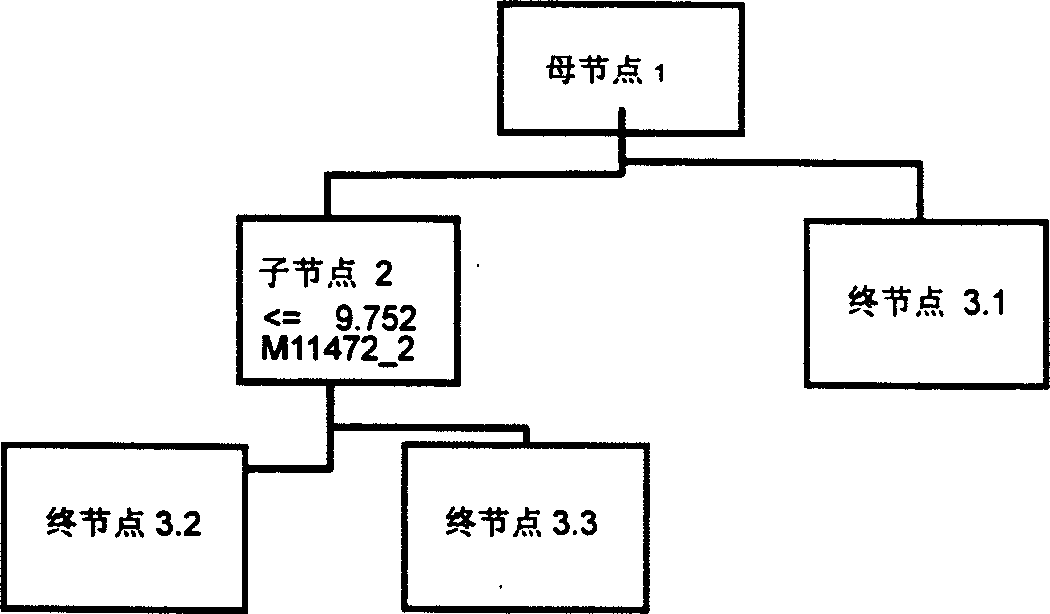
Mass spectrum model for detecting liver cancer serum characteristic protein and method for preparation
A technology of characteristic protein and mass spectrometry model, which is applied in the field of protein fingerprint detection to achieve high sensitivity and specificity, reasonable and feasible construction method, and the effect of reducing the fatality rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Draw 2 mL of venous blood from the patient to be tested without anticoagulation. Place at room temperature for 30 min to 1 h, centrifuge at room temperature at 2500 rpm for 5 min, take aliquots of serum and store in a -70°C refrigerator. During the experiment, the samples were taken out from the -70°C refrigerator, thawed in ice, and centrifuged at 10,000 rpm for 2min at 4°C. Take 3 μL of centrifuged serum sample, add 6 μL of U9 treatment solution (9M urea, 2% CHAPS, 1% DTT, 50mM Tris-CL, pH9.0), mix well, shake in ice bath for 30min, take it out, add 108μl of binding buffer (100mmol / LNaAc, PH4.0), mix immediately. Put the WCX2 chip into the bioprocessor, add 200 μl of binding buffer to each well, shake and wash at room temperature for 2 times, each time for 5 minutes, and spin dry. Add 100 μl sample mixture to each well, shake and incubate for 1 h, shake off the sample, wash twice with 200 μl elution buffer (100 mmol / L NaAc, pH 4.0) at room temperature for 5 min each...
Embodiment 2
[0046] Draw 2 mL of venous blood from the patient to be tested without anticoagulation. Place at room temperature for 30 min to 1 h, centrifuge at room temperature at 2500 rpm for 5 min, take aliquots of serum and store in a -70°C refrigerator. During the experiment, the samples were taken out from the -70°C refrigerator, thawed in ice, and centrifuged at 10,000 rpm for 2min at 4°C. Take 3 μL of centrifuged serum sample, add 6 μL of U9 treatment solution (9M urea, 2% CHAPS, 1% DTT, 50mM Tris-CL, pH9.0), mix well, shake in ice bath for 30min, take it out, add 108μl of binding buffer (100mmol / LNaAc, PH4.0), mix immediately. Put the WCX2 chip into the bioprocessor, add 200 μl of binding buffer to each well, shake and wash at room temperature for 2 times, each time for 5 minutes, and spin dry. Add 100 μl sample mixture to each well, shake and incubate for 1 h, shake off the sample, wash twice with 200 μl elution buffer (100 mmol / L NaAc, pH 4.0) at room temperature for 5 min each...
Embodiment 3
[0048] Draw 2 mL of venous blood from the patient to be tested without anticoagulation. Incubate at 37°C for 20-30min, centrifuge at room temperature at 2500rpm for 5min, take aliquots of serum and store in -70°C refrigerator. During the experiment, the samples were taken out from the -70°C refrigerator, thawed in ice, and centrifuged at 10,000 rpm for 2min at 4°C. Take 3 μL of centrifuged serum sample, add 6 μL of U9 treatment solution (9M urea, 2% CHAPS, 1% DTT, 50mM Tris-CL, pH9.0), mix well, shake in ice bath for 30min, take it out, add 108μl of binding buffer (100mmol / LNaAc, PH4.0), mix immediately. Put the WCX2 chip into the bioprocessor, add 200 μl of binding buffer to each well, shake and wash at room temperature for 2 times, each time for 5 minutes, and spin dry. Add 100 μl sample mixture to each well, shake and incubate for 1 h, shake off the sample, wash twice with 200 μl elution buffer (100 mmol / L NaAc, pH 4.0) at room temperature for 5 min each time, and dry; th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com