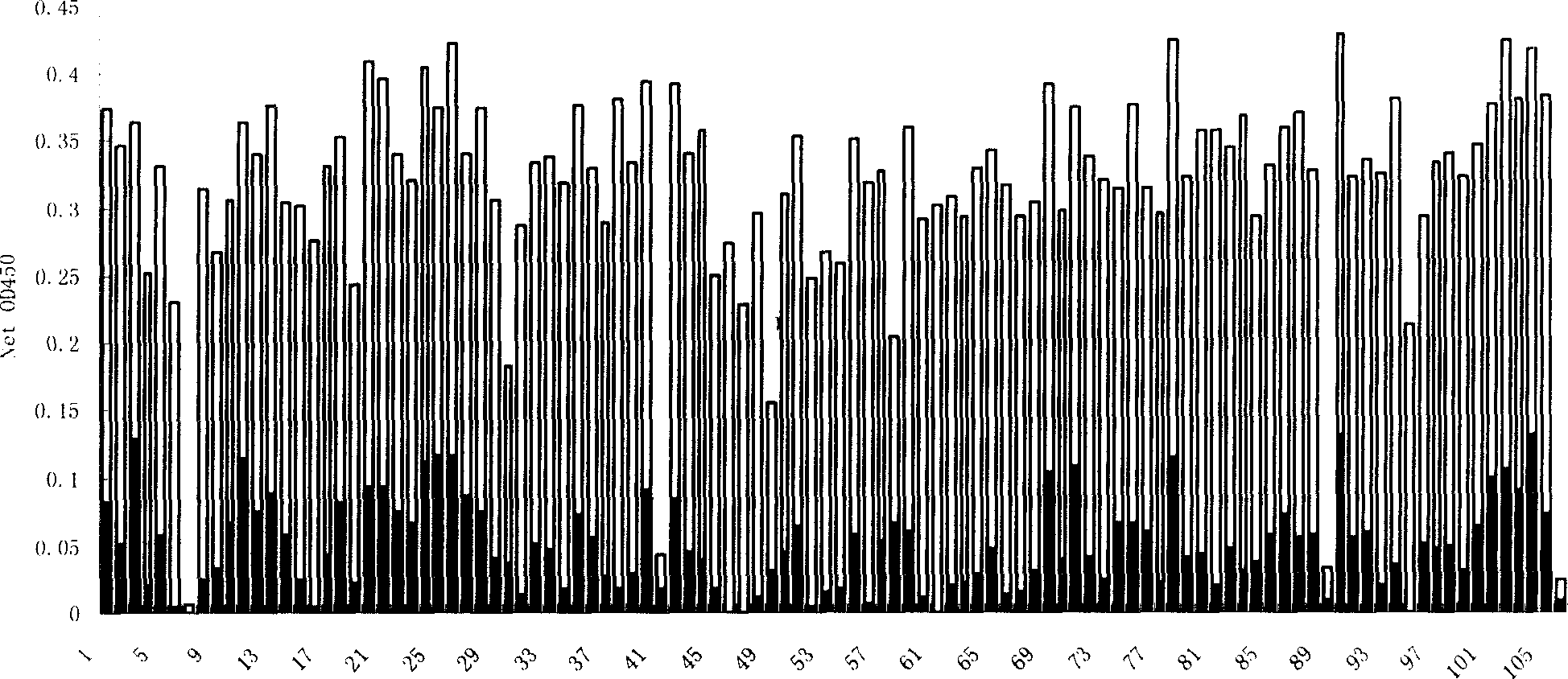Screening and authentication of polypeptide binding specificly to tumour tranferring cell
A tumor metastasis and specific technology, applied in peptides, material inspection products, biological tests, etc., can solve the problems of complex, unknown and non-specific components on the cell surface
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
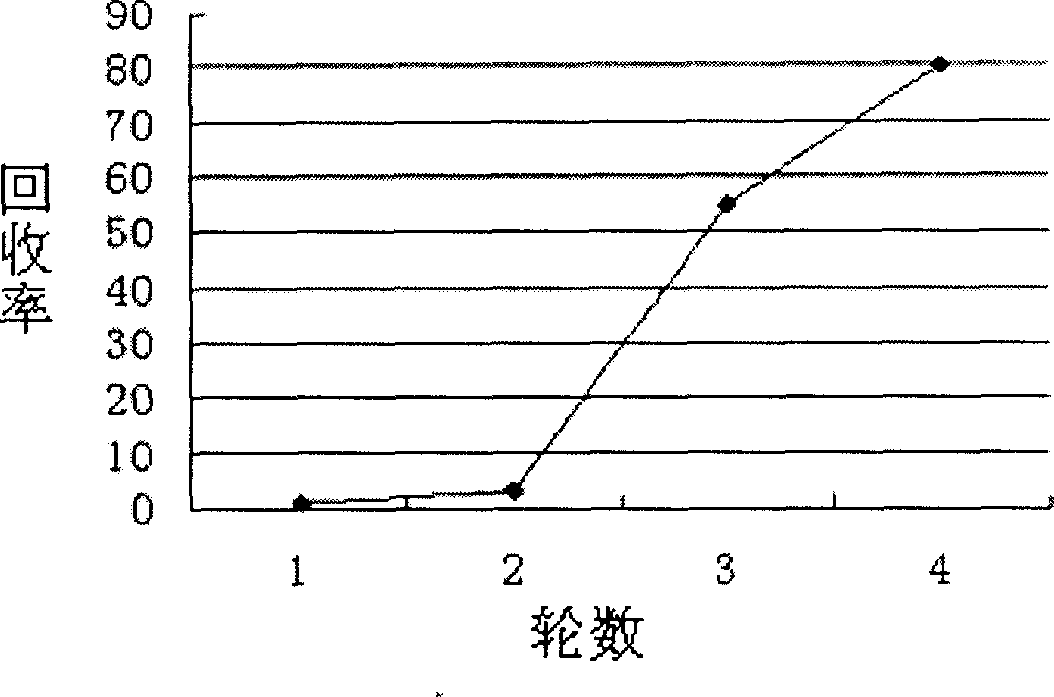
[0028] Example 1: Obtaining recombinant phage capable of selectively binding to the surface of SW620 cells after four rounds of screening
[0029] The first round: Dissolve 10.4g of RPMI1640 medium powder (Gibico company) and 2.0g of sodium bicarbonate in 800ml of double distilled water, add 10ml of 1mol / L HEPES solution, and then distill the total volume to 1000ml with double distilled water, 0.22 Store at 4°C after sterilizing with a μm filter membrane; add fetal bovine serum (FBS, purchased from Tianjin Institute of Blood Diseases) and penicillin-streptomycin double antibody to the above medium to make the content reach 10% and 1% respectively , stored at 4°C; 2ml of the above-mentioned medium was added to each well of a 6-well plate, and SW620 cells (ATCC deposit number: CCL-228) were inserted to make the cell density 10 6 cells / well, cultivated in a carbon dioxide cell incubator for about 48 hours; discard the medium, and wash the cells with 1×PBS (10×PBS: sodium chloride...
Embodiment 2
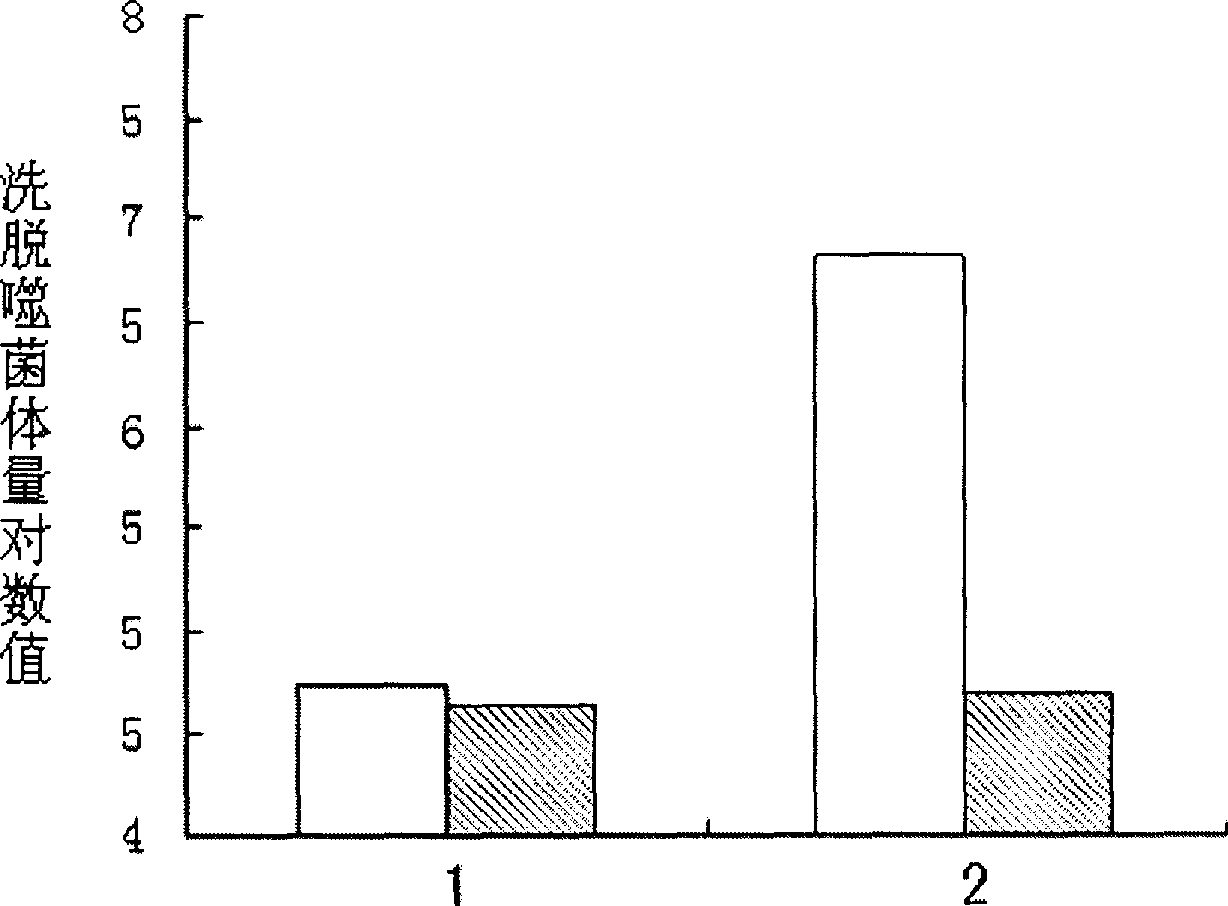
[0037] Example 2: Determination of phage binding ability to SW480 and SW620
[0038] SW480 and SW620 respectively with 10 6 Spread the cells / well into a 6-well plate, culture in a carbon dioxide cell incubator at 37°C for about 48 hours (the medium uses RPMI1640, 10% FBS, 1% penicillin-streptomycin double antibody), discard the medium, and wash with PBS , add blocking solution (PBS containing 10mg / ml BSA) and incubate at room temperature for 1 hour; at the same time block the phage, discard the blocking solution, and add 2ml of phage to each of the SW480 and SW620 wells (the titer is about 1×10 11 ), incubated at room temperature for 2 hours; discarded the phage, washed 10 times with PBST (PBS containing 0.1% Tween), washed twice with PBS, eluted, and measured their respective titers: the original library phage was used as a control, and the above-mentioned Test; the phage mixed library obtained in the fourth round has a binding ability to SW620 that is 100 times higher than ...
Embodiment 3
[0043] Example 3: Isolation and preparation of monoclonal phage
[0044] Randomly pick 105 monoclonal phages after 4 rounds of screening, add them to Escherichia coli ER2738 in mid-logarithmic phase, and culture them on a shaker (250 rpm) at 37°C for 4.5 to 5 hours; centrifuge the amplified product at 10,000 rpm For 10 minutes, take the supernatant and add 1 / 6 volume of PEG / NaCl solution, precipitate overnight at 4°C; centrifuge at 10,000 rpm for 15 minutes, discard the supernatant: resuspend the pellet in 200 μl of PBS, and centrifuge at 10,000 rpm for 5 minutes to remove the remaining Bacterial precipitation; measure the phage titer in the supernatant by agar stacking method, add an equal volume of glycerol, and store at -20°C.
[0045] Complete cell SW480 or SW620 was detected by ELISA with 105 monoclonal phages. Into a 96-well plate, add 10 5 For SW480 or SW620 cells / well, add 200 μl of blocking solution [5% (w / v) BSA in PBS] to each well after two days of culture, and b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com