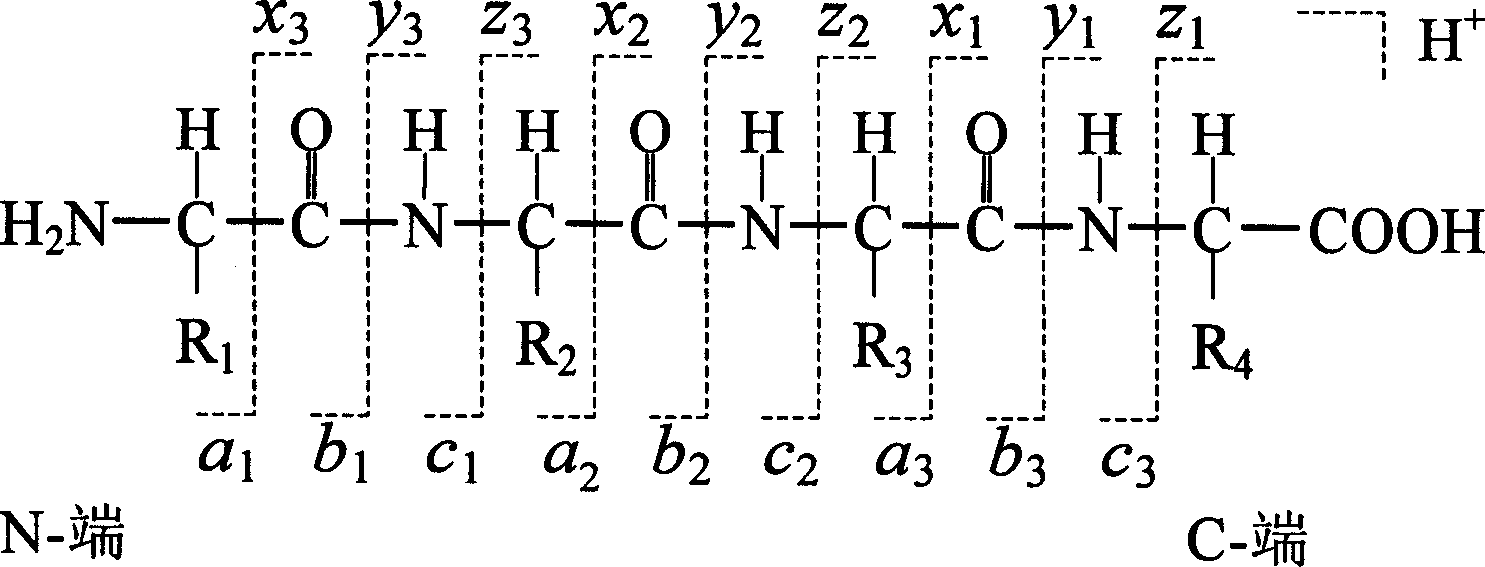Method for identifying peptide by using tandem mass spectrometry data
A tandem mass spectrometry and data identification technology, applied in the field of proteome analysis, can solve the problems of unpredictable peptide fragmentation mode and wrong peptide identification, and achieve the effect of reducing false positive results and high accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] The present invention will be further described in detail below in conjunction with the accompanying drawings and specific embodiments.
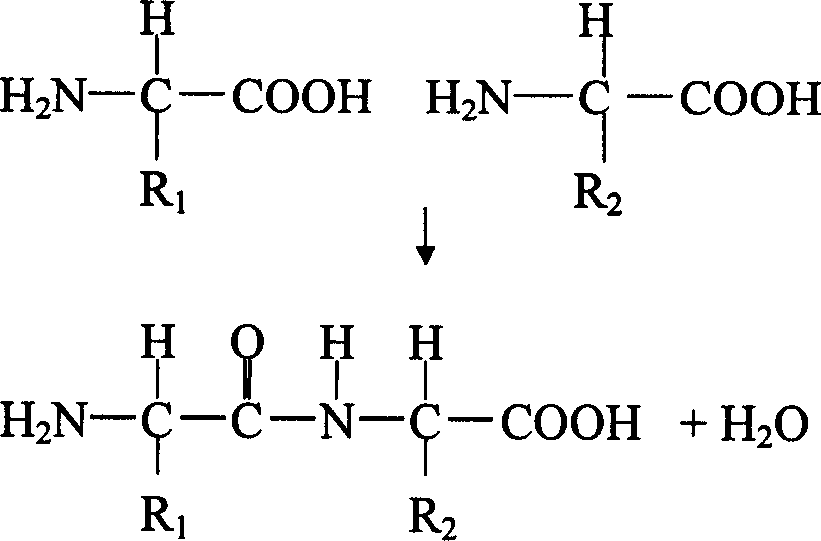
[0027] Such as figure 1 As shown, two amino acids can be connected by losing a water molecule to form a peptide bond at their C-terminal and N-terminal. A peptide is a sequence of amino acid residues connected to each other by peptide bonds. This sequence determines the identity of the peptide.
[0028] To identify the amino acid sequence of a peptide, the peptide is ionized and then passed into a mass spectrometer. In a mass spectrometer, peptide ions with a specific mass-to-charge ratio (m / z) (these peptide ions usually have the same amino acid sequence) are fragmented under the action of Collision-Induced Dissociation (CID). Under the action of low-energy CID, the rib bond can usually be broken in three ways to generate six series of fragment ions, namely a, b, c at the N-terminal and x, y, z series of fragment ions at the C-term...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com