Common protein surface shapes and uses therefor
a protein surface and surface technology, applied in the field of identifying and representing the common three-dimensional structural features of proteins, can solve the problems of not being able to identify molecules that could modulate biological functions, unable to know which descriptors are important or essential, and unable to describe the structural elements of various molecular recognition events
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
1 The Clustering of β-Turns
1.1 Background
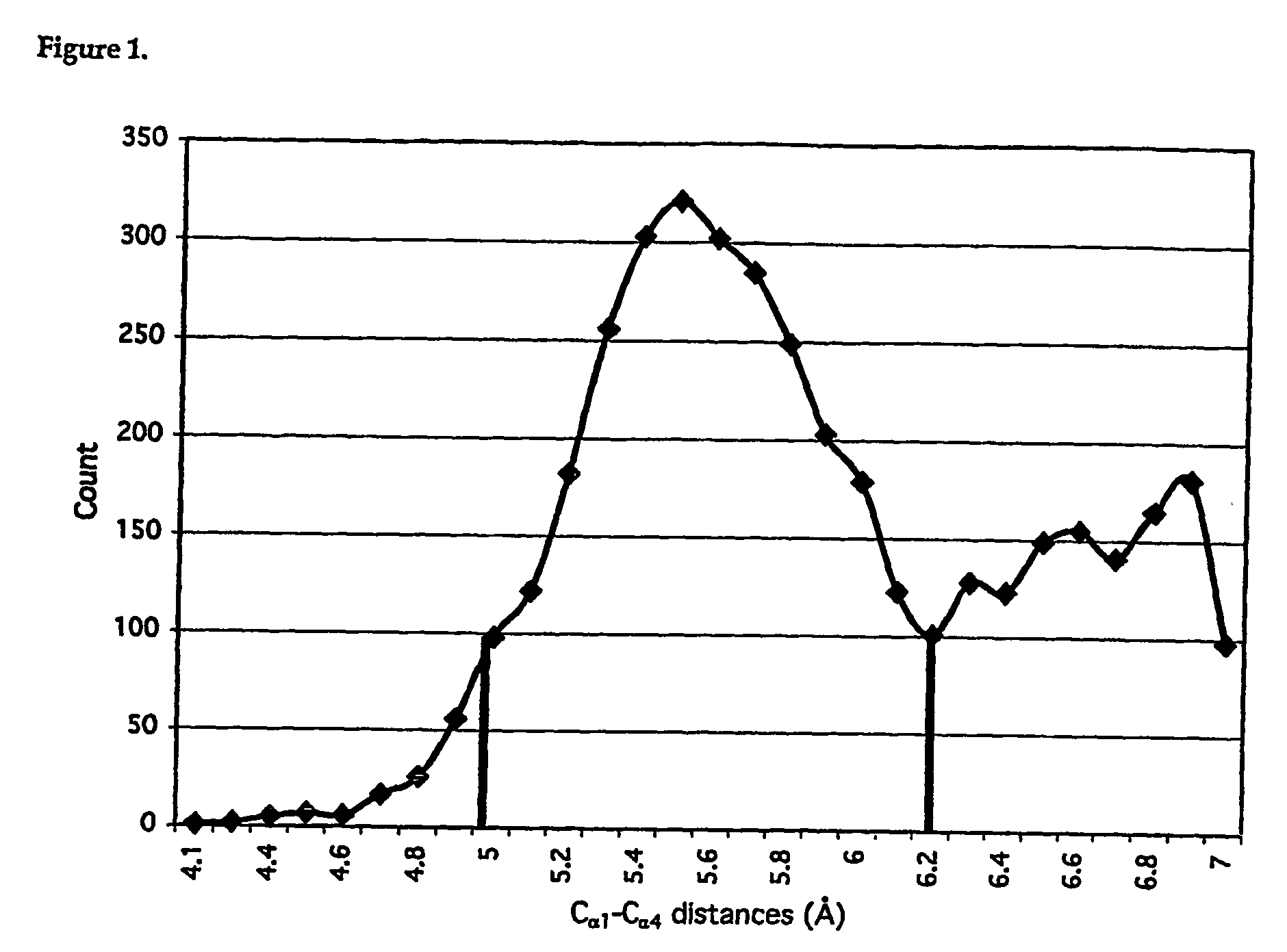
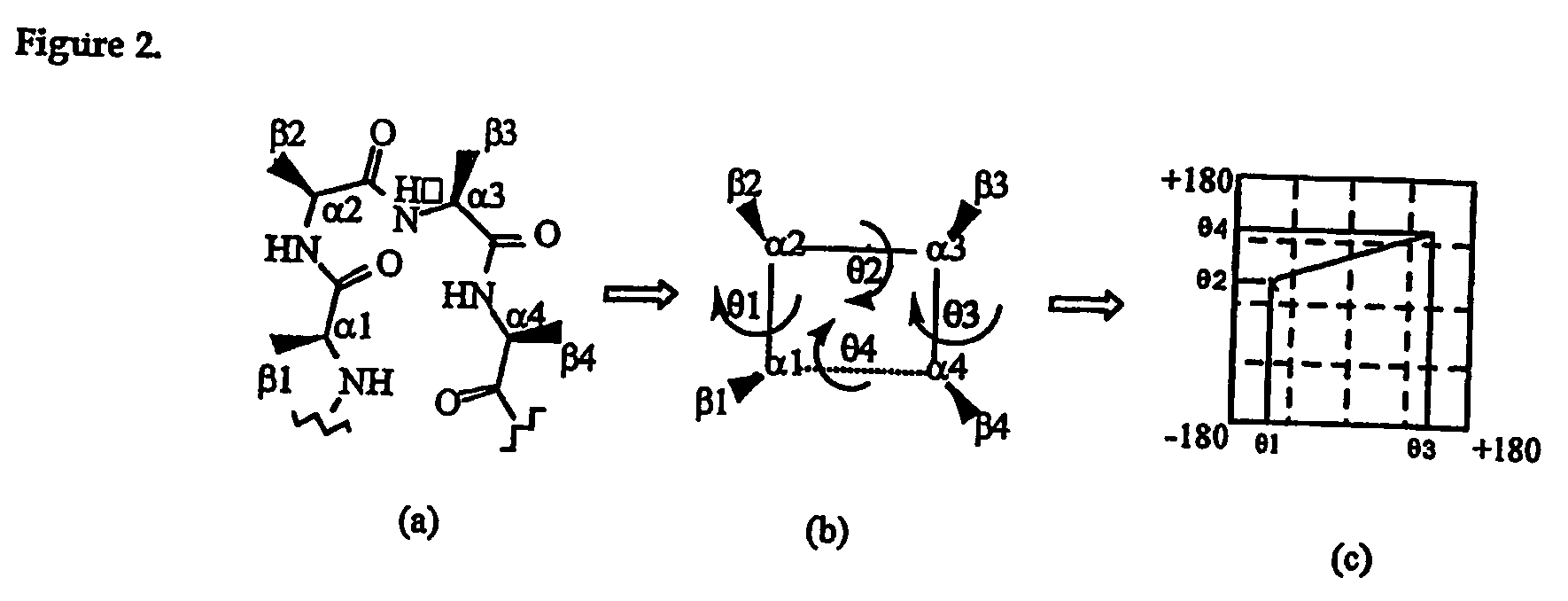
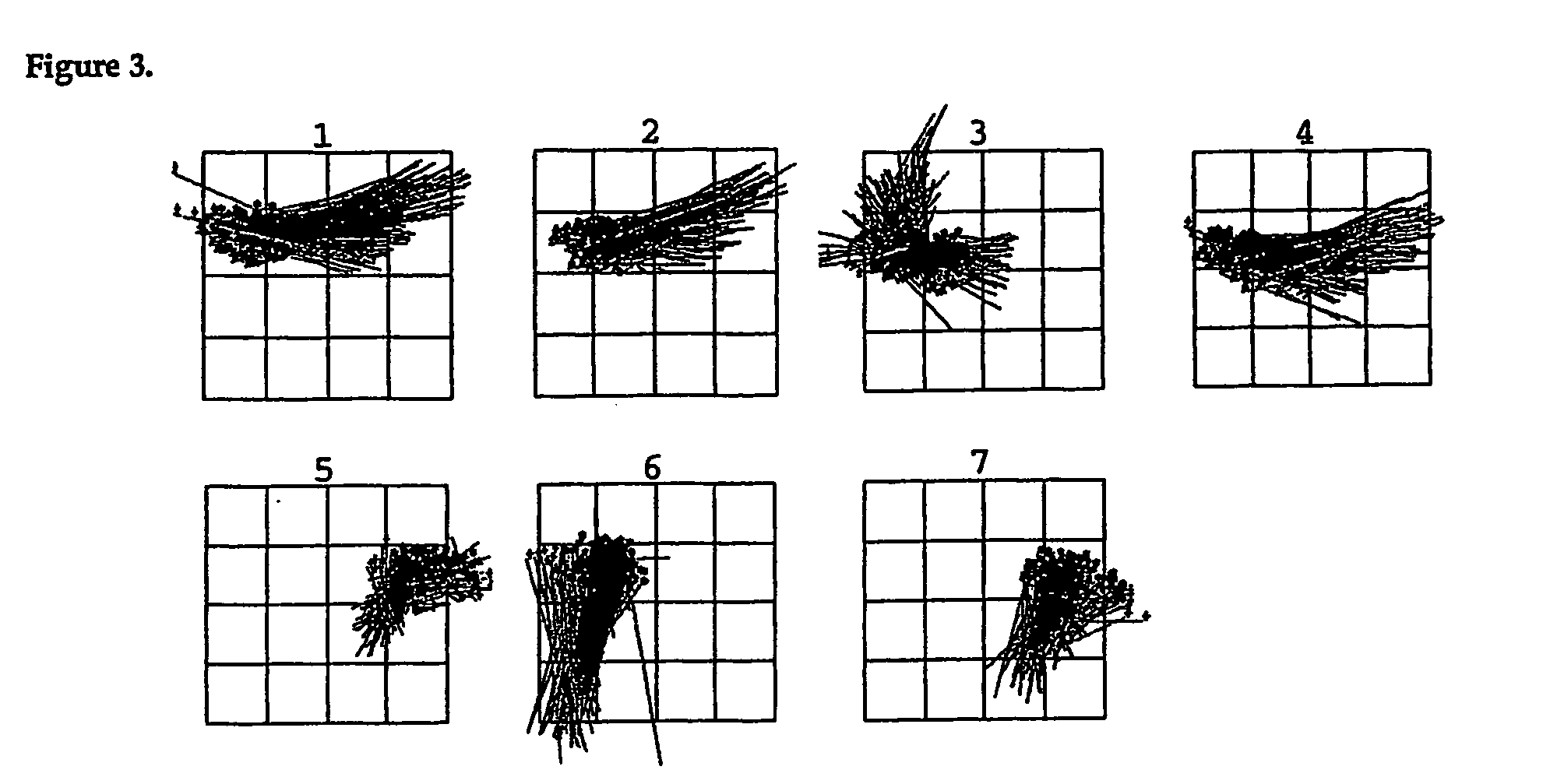
[0151] Protein structure comprises stretches of secondary structure (helices or β-sheets) that are joined by turns, which enable a reversal in chain direction. These turns are normally positioned on the surfaces of proteins and allow the formation of the globular protein interior1. β-turns2-4 are more common than the tighter coiled γ-turns and the looser coiled α-turns and have been defined as four residue segments of polypeptides in which the distance between cαi and cαi+3 is less than 7 Å, and that the central residues are not helical5. β-turns encompass 25% of residues in proteins6, are important for protein and peptide function2,7-9, and are an important driving force in protein foldinge2,10,11. Consequently, there have been numerous studies on the design and development of β-turn mimetics7,12-22.
[0152] Despite the importance of side chain spatial arrangement in molecular recognition, the conformations of β-turns are currently classi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| shape | aaaaa | aaaaa |
| surface shape | aaaaa | aaaaa |
| surface charge shape | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com