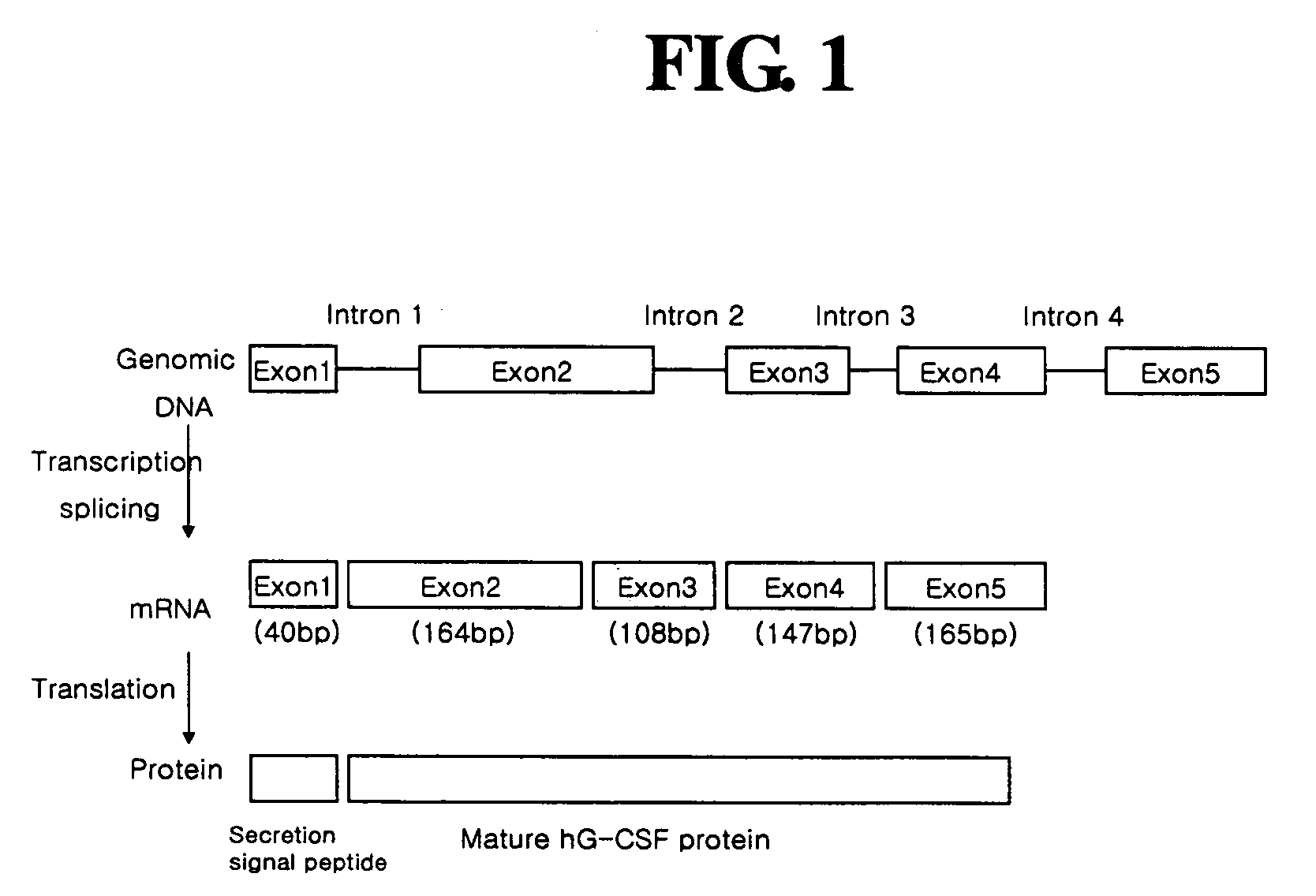
Diagnostic method for cancer characterized in the detection of the deletion of G-CSF exon 3
a cancer and exon technology, applied in the field of cancer diagnosis, can solve the problems of generating false positive data, morphological and immunohistochemical diagnosis requires much longer time, and high cost, and achieves the effect of simple and rapid diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of mRNA and cDNA from Tumor Cell Lines
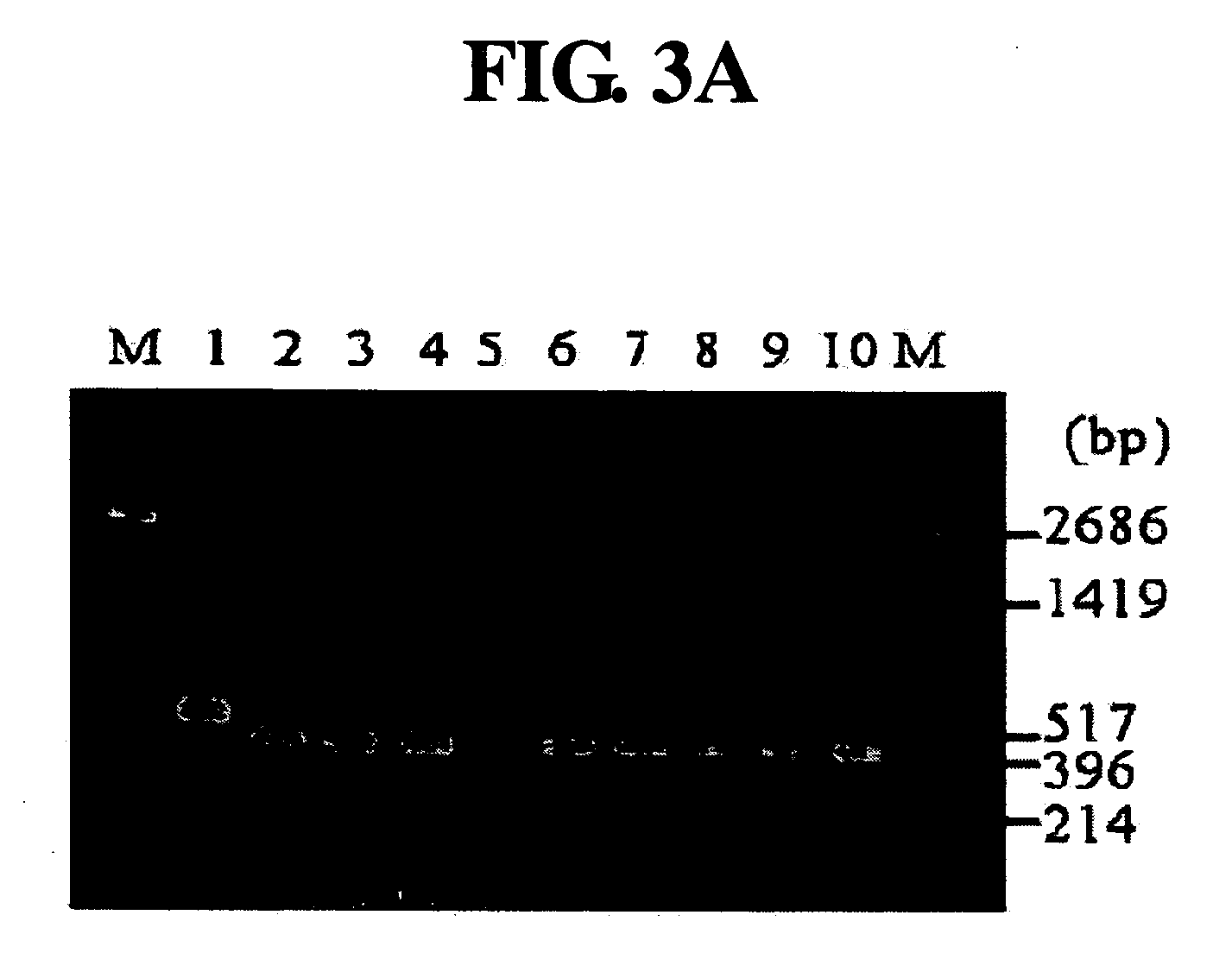
[0055] mRNA and cDNA samples were prepared from 8 normal cell lines and tissues, and 17 tumor cell lines. The normal cell lines and tumor cell lines used in Examples of the present invention are given in Table 1, below.
Table 1
Normal and Tumor Cell Lines Used in the Present Invention
[0056]
Cell typesCell collection centersTumor cellYCC-7Stomach cancer cell lineCancer metastasis research center,linesCollege of Medicine, Yonsei UniversityAGSStomach cancer cell lineATCC CRL-1739SNU-1Stomach cancer cell lineKorean Cell Line Research Foundation(KCLRF), Seoul National UniversityMDA-MB-Breast cancer cell lineATCC HTB-26231MCF-7Breast cancer cell lineATCC HTB-22SK-BR-3Breast cancer cell lineATCC HTB-30HT-1080Sarcoma cell lineATCC CCL-121HCT-116Colon cancer cell lineATCC CCL-247COLO205Colon cancer cell lineATCC CCL-222DLD-1Colon cancer cell lineATCC CCL-221HT-29Colon cancer cell lineATCC HTB-38A549Lung cancer cell lineATCC CCL-185NCI-H460Lu...
example 2
Detection of hG-CSF Gene by PCR
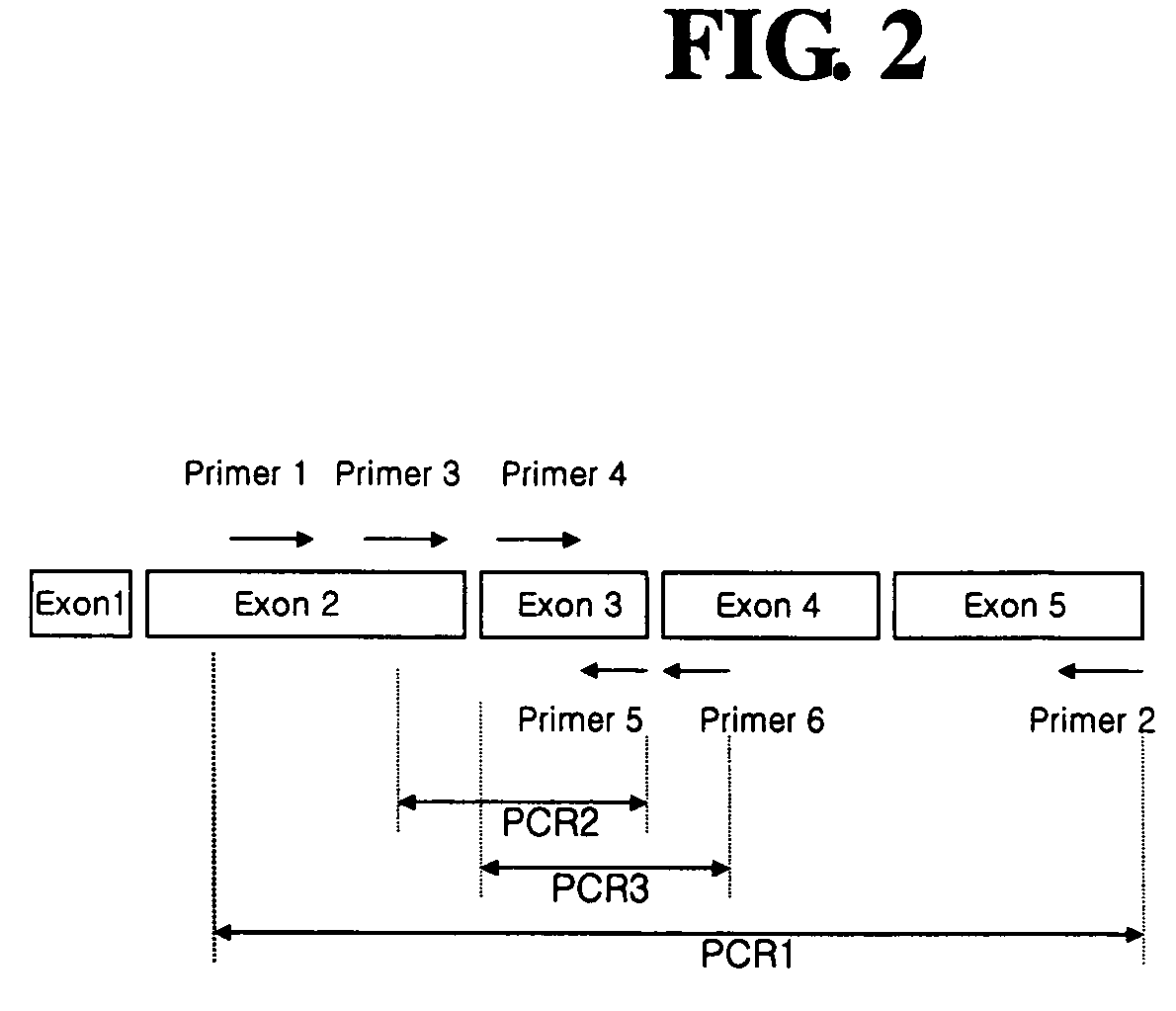
[0060] In order to detect expression of normal hG-CSF gene in each tumor cell line, PCR was carried out using cDNA prepared in Example 1 as a template. As shown in FIG. 2, PCR reactions were divided into three types according to their amplified products, as follows: PCR 1 for amplification of a region (Thr1-Pro174) ranging from a part of exon 2 to exon 5 of hG-CSF gene; PCR 2 for amplification of a region (Ile24-Leu71) ranging from a part of exon 2 to exon 3 of hG-CSF gene; and PCR 3 for amplification of a region (Cys36-Ser80) ranging from exon 3 to a part of exon 4 of hG-CSF gene.
[0061] PCR 1 was carried out using a cDNA sample from each tumor cell line as a template, and a primer set of a sense primer designated SEQ ID NO.: 1 (5′-ACCCCCCTGGGCCCTGCC-3′) and an antisense primer designated SEQ ID NO.: 2 (5′-TCAGGGCTGGGCAAGGTG-3′). PCR 2 was carried out using a cDNA sample from each tumor cell line as a template, and a primer set of a sense primer desi...
example 3
Detection of G-CSF Gene by Hybridization
[0065] The deletion of exon 3 in G-CSF cDNA from tumor cell lines was detected by hybridization.
[0066] PCR was carried out using G-CSF gene derived from a normal cell line as a template, and a primer set of a sense primer designated SEQ ID NO.: 4 (5′-TGTGCCACCTACAAGCTGTGC-3′) and an antisense primer designated SEQ ID NO.: 5 (5′-CAGCTGCAGGGCCTGGCT-3′). A DNA fragment of 108 bp corresponding to the exon 3 region of G-CSF gene was obtained.
[0067] Separately, PCR was carried out using G-CSF gene derived from the normal cell line as a template, and a primer set of a sense primer designated SEQ ID NO.: 1 (5′-ACCCCCCTGGGCCCTGCC-3′) and an antisense primer designated SEQ ID NO.: 9 (5′-CAGCTTCTCCTGGAGCGC-3′). A DNA fragment of 105 bp corresponding to the exon 2 region of G-CSF gene was obtained.
[0068] After being purified, each of the DNA fragments (50 ng / μl) was spotted on a nylon membrane (Boehringer Mannheim, Germany), and incubated at 80° C. fo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com