Three-dimensional structure of complement receptor type 2 and uses thereof
a technology of complement receptor and three-dimensional structure, which is applied in the field of three-dimensional structure of complement receptor type 2, can solve the problems of entanglement of the molecular recognition mechanism and achieve the effect of improving the molecular recognition efficiency and sensitivity of this class of proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
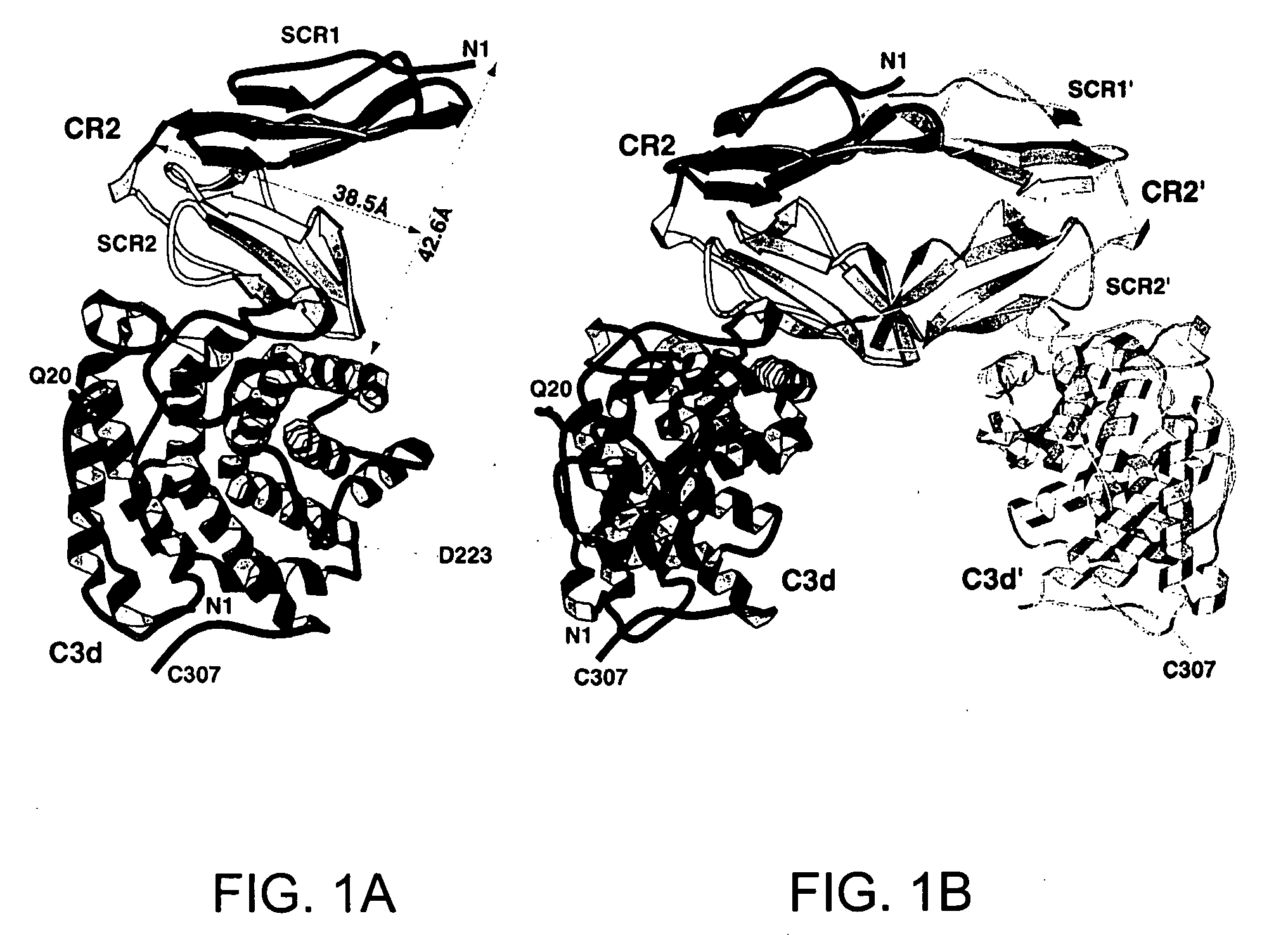
[0161] The following example describes the crystallization and structure determination of the complex of complement receptor type 2 (CR2) and C3d.
Crystallization and Structure Determination of Structure
[0162] The crystals of the complex of CR2-C3d were obtained by co-crystallization of CR2 and C3d, at a protein ratio where no free CR2 or C3d could be detected by native gel electrophoresis. The protein concentration of 20 mg / ml was used for crystallization by the method of hanging drop vapor diffusion. The crystallization buffer contained 17% PEG 2K, 0.2 M ZnAcetate, and 0.1 M NaCacodylate (pH 7.36). Crystals reached full size after 4-6 weeks at 4° C. Crystal was frozen under liquid nitrogen in the crystallization buffer containing 20% glycerol. Synchrotron data were collected at Brookhaven National Laboratory and was indexed, integrated and reduced using D*trek (licensed through MSC Inc., Table 1). The space group is R32, with unit cell a=b=170.5 Å, c=173.8 Å. AmoRe (CCP4, Acta C...
example 2
[0183] The following example describes the construction of C3d mutants that affect CR2 binding.
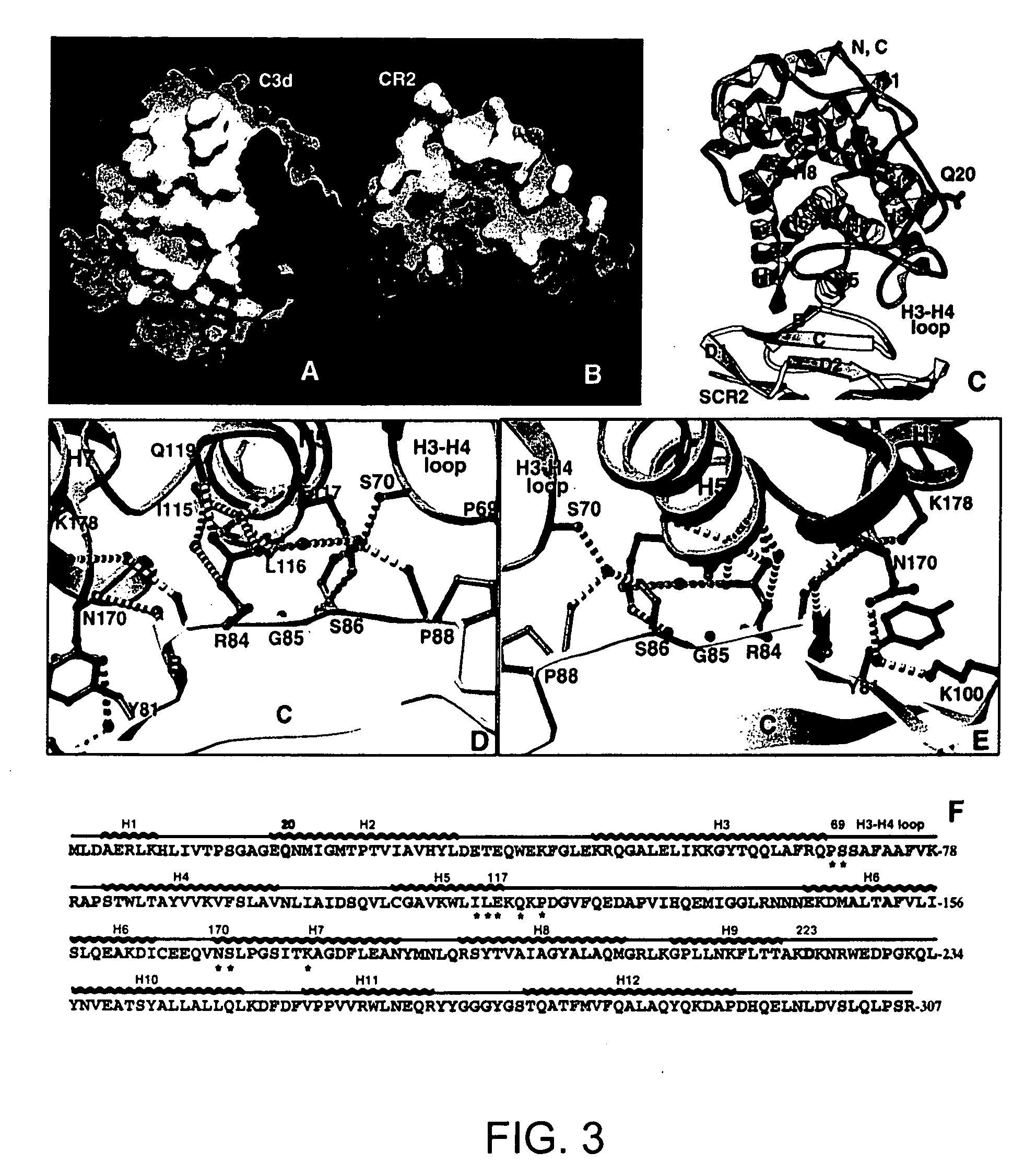
[0184] Based on the complex structure, mutagenesis of C3d around the interface to disrupt CR2 binding was predicted to be difficult. This is because the interaction between CR2 and C3d involves mostly main-chain H-bonding, and the side chain residues play relatively small roles in the binding. However, to confirm the accuracy of the CR2-C3d interaction seen in this co-crystal structure, two informative C3d mutants were constructed. In mutant 170 (mt170; SEQ ID NO:8), residue Asn170 was changed to Arg. Asn170 is located on H7 of C3d and is the only residue on C3d that more or less points directly toward CR2 in the interface (FIG. 3E). Asn170 also packs directly with Tyr81 of CR2 as well as forms an H-bond with CR2 (Lys 100) through a water molecule (FIG. 3E). In solution, this mutant protein behaved very similarly like the wild type C3d and showed the same apparent molecular weight as the ...
example 3
[0190] The following example describes the inhibition of CR2-C3d interaction by CR2-derived peptides.
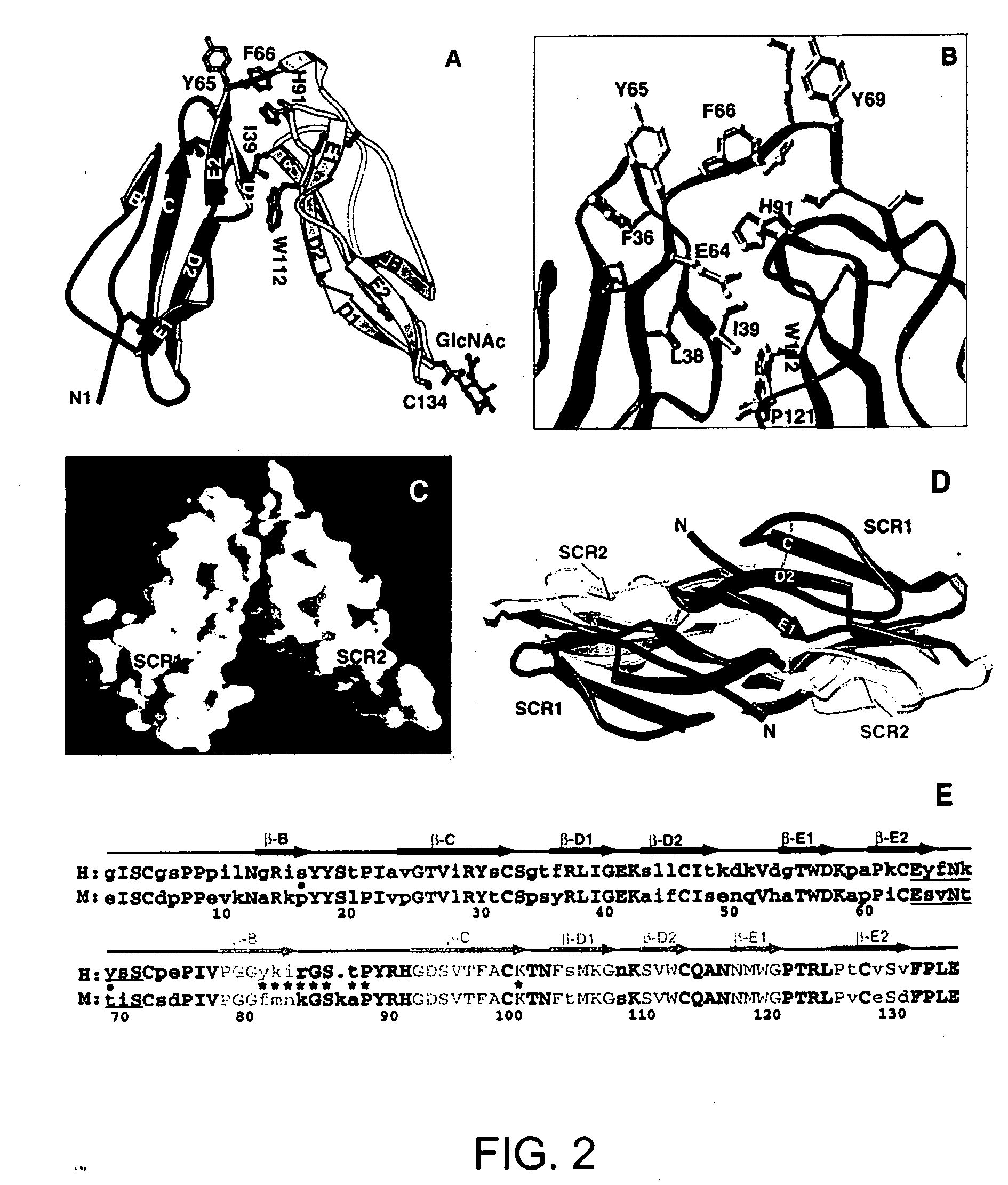
[0191] Based on the structure of the CR2 L-C3d complex, the results from previously reported CR2 peptide inhibition and monoclonal antibody assays can now be explained. In the peptide inhibition tests using short synthetic peptides covering all of CR2 SCR1 and SCM, peptides from two regions were shown to inhibit CR2-C3 binding (H. Molina, et al., J Immunol 154,5426-35 (1995)). One of them contains sequences that are located right on the interaction interface of CR2 seen in the complex structure, namely the sequences from the B strand and B-C loop of SCR2 (FIGS. 3D & 3E). This independent result strongly supports the complex structure described by the present invention. However, the other peptide located on the B strand of SCR1 also showed a similar inhibition effect as the first one. Close examination of this fragment on CR2 structure within the SCR1 domain revealed a very similar 3...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com