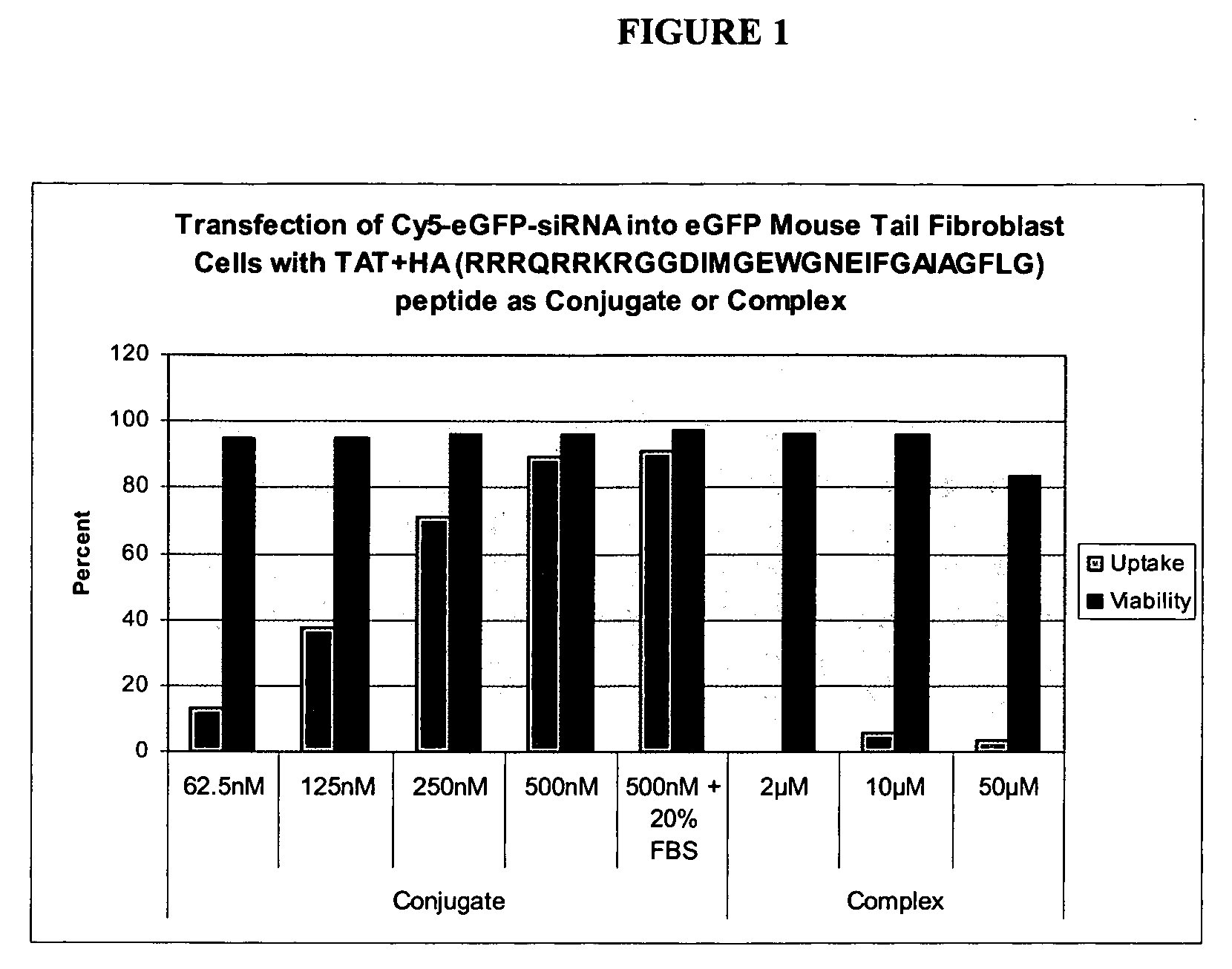
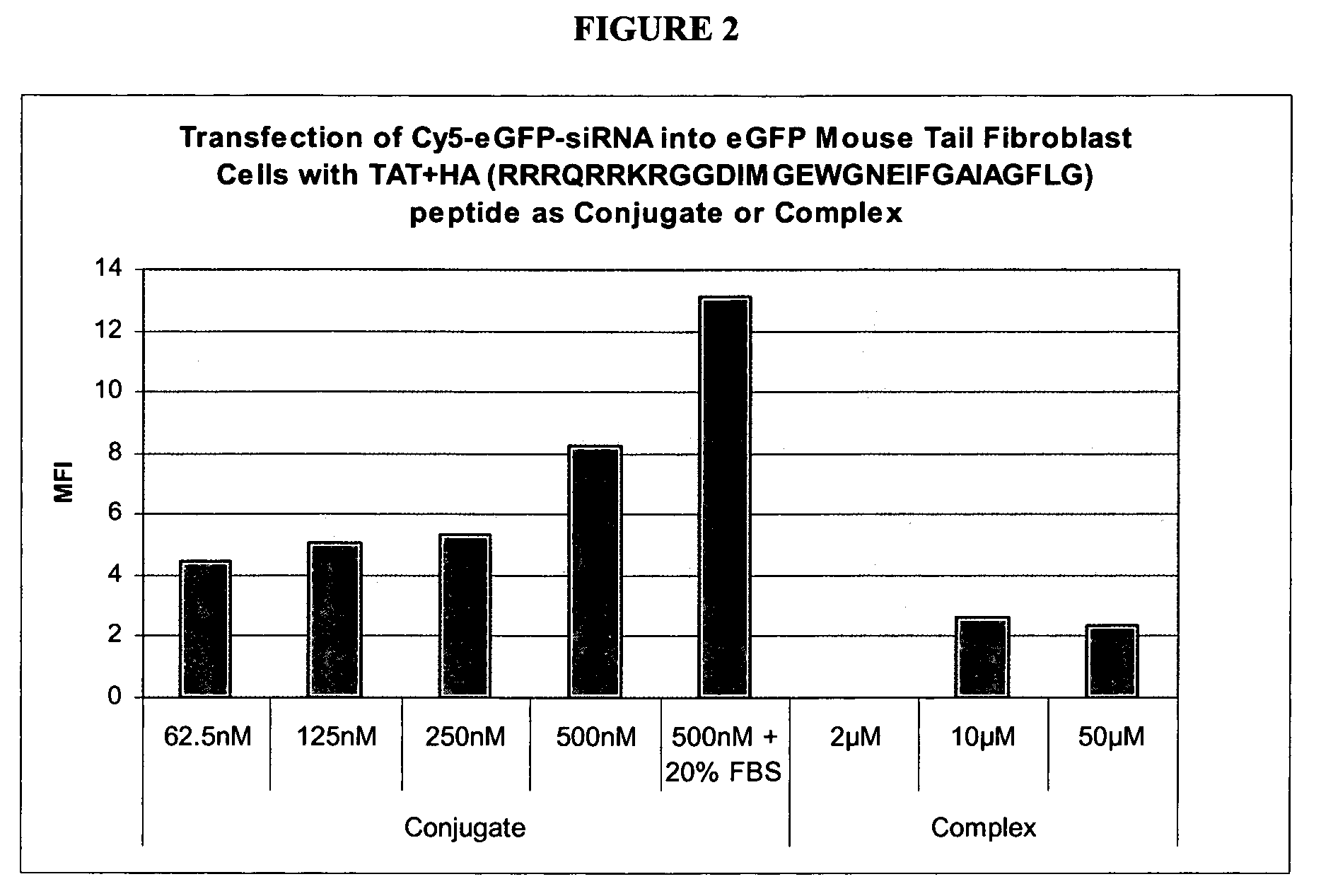
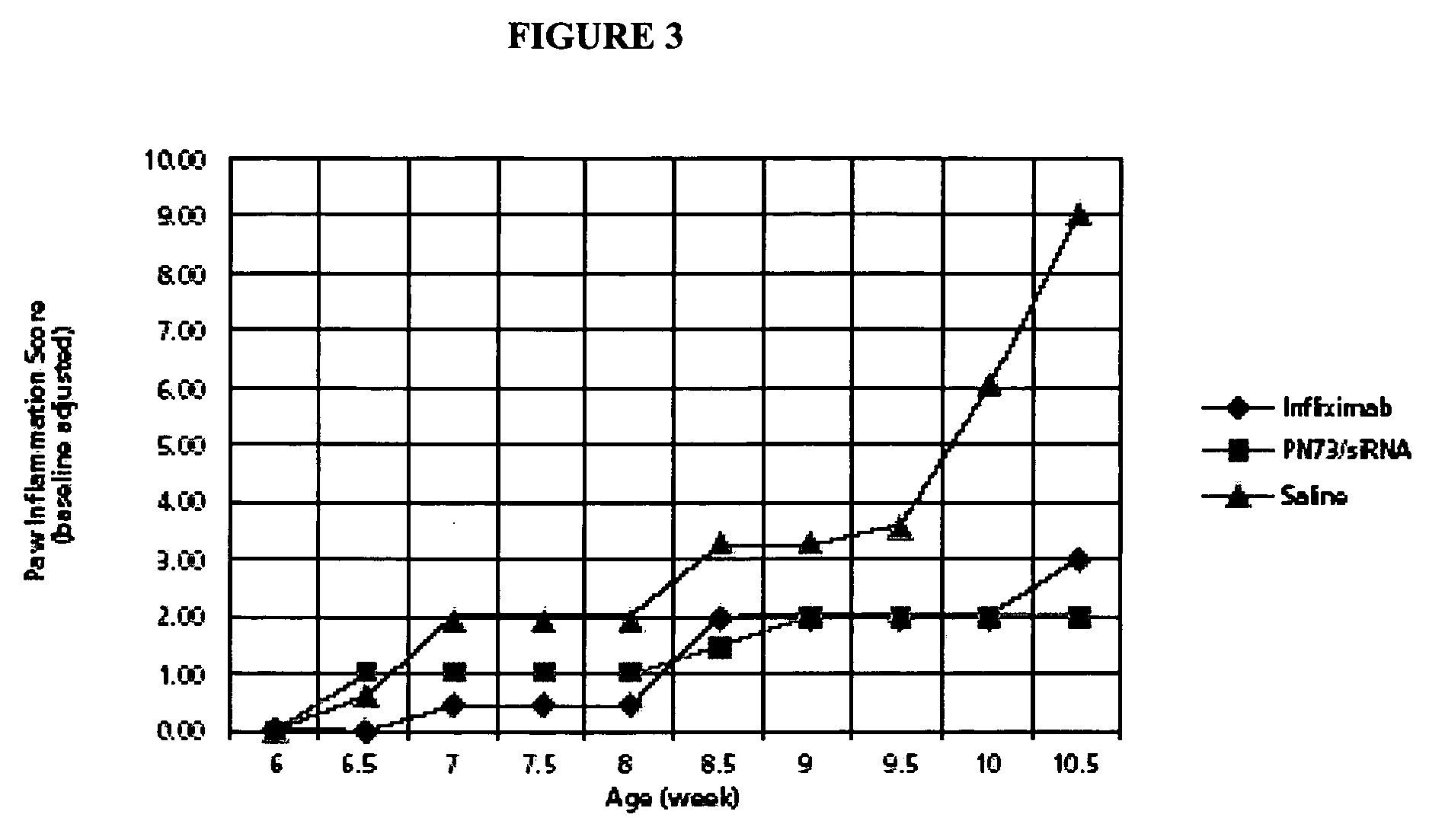
Compostions and methods for enhancing delivery of nucleic acids into cells and for modifying expression of target genes in cells
a nucleic acid and cell technology, applied in the field of nucleic acid delivery into cells, can solve the problems of limited nucleic acid delivery techniques, poor efficiency, toxicity of delivery reagents,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Production and Characterization of Compositions Comprising a siRNA Complexed with a Polynucleotide Delivery-Enhancing Polypeptide
[0132] To form complexes between candidate siRNAs and polynucleotide delivery-enhancing polypeptides of the invention, an adequate amount of siRNA is combined with a pre-determined amount of polynucleotide delivery-enhancing polypeptide, for example in Opti-MEM® cell medium (Invitrogen), in defined ratios and incubated at room temperature for about 10-30 min. Subsequently a selected volume, e.g., about 50 μl, of this mixture is brought into contact with target cells and the cells are incubated for a predetermined incubation period, which in the present example was about 2 hr. The siNA / peptide mixture can optionally include cell culture medium or other additives such as fetal bovine serum. For H3, H4 and H2b, a series of experiments was performed to complex these polynucleotide delivery-enhancing polypeptides with siRNA in different ratios. Generally this ...
example 2
Production and Characterization of Compositions Comprising a siRNA Comjugated With a TAT-HA Polynucleotide Delivery-Enhancing Polypeptide
[0171] The present example describes the synthesis and uptake activity of specific peptides covalently conjugated to one strand of a siRNA duplex. These conjugates efficiently deliver siRNA into the cytoplasm and mediate knockdown of desired target genes.
Peptide Synthesis
[0172] Peptides were synthesized by solid-phase Fmoc chemistry on CLEAR-amide resin using a Rainin Symphony synthesizer. Coupling steps were performed using 5 equivalents of HCTU and Fmoc amino acid with an excess of N-methylmorpholine for 40 minutes. Fmoc removal was accomplished by treating the peptide resin with 20% piperidine in DMF for two 10 minutes cycles. Upon completion of the entire peptide, the Fmoc group was removed with piperidine and washed extensively with DMF. Maleimido modified peptides were prepared by coupling 3.0 equivalents of 3-maleimidopropionic acid and ...
example 3
Screening of siRNA / Delivery Peptide Complexes Demonstrates Efficient Induction of siRNA Uptake in 9L / LacZ Cells by a Diverse Assemblage of Rationally-Designed Polynucleotide Delivery-Enhancing Polypeptides
[0179] The present example provides additional evidence that a broad and diverse assemblage of rationally-designed polynucleotide delivery-enhancing polypeptides of the invention induce or enhance siRNA uptake when complexed with siRNAs
[0180] Approximately 10,000 9L / lacZ cells were plated per well in flat-bottom 96-well plates so that they would be ˜50% confluent the next day at the time of transfection. FAM-labeled siRNA and peptides were diluted in Opti-MEM® media (Invitrogen) at 2-fold the final concentration. Equal volumes of siRNA and peptide were mixed and allowed to complex 5-10 minutes at room temperature and then 50 μL added to cells, previously washed with PBS. Cells were transfected for 3 hours at 37° C., 5% CO2. Cells were washed with PBS, treated with trypsin and the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Dimensionless property | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com