Fgf2-related methods for diagnosing and treating depression
a depression and fgf2 technology, applied in the field of fgf2related methods for diagnosing and treating depression, can solve the problems of ineffective and reliable diagnosis and rate, lack of biomarkers, and difficulty in diagnosing bipolar disorder, and achieve the effect of accurate diagnosis of mental illness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
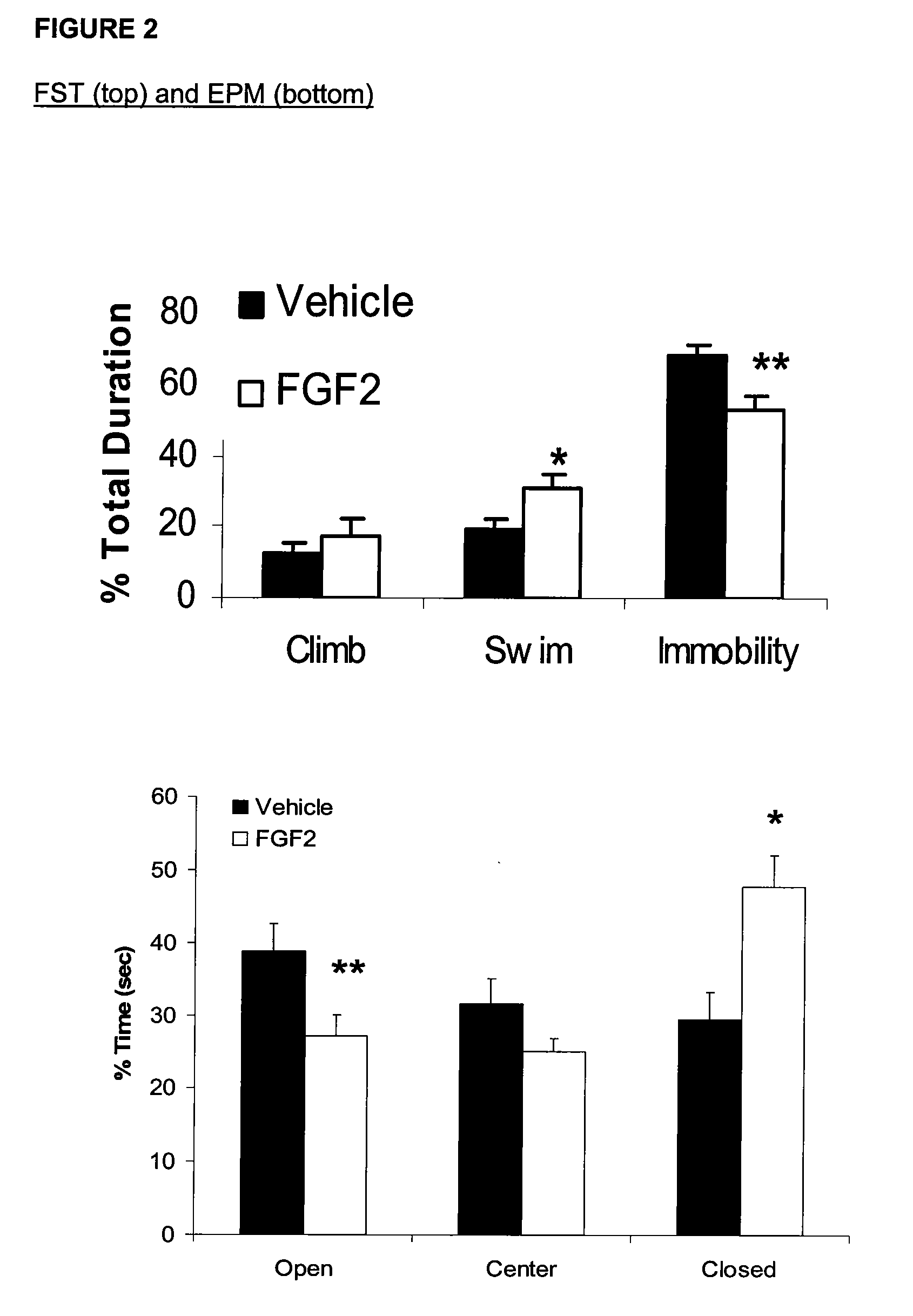
Differential Expression of Genes Associated with Suicide in Both BP and MDD Subjects
[0257]Previous studies have investigated genes associated with mood disorders and suicidal tendencies, using microarrays and PCRs to analyze gene expression (Sibille et al., 2004; Yanagi M, et al., J Hum Genet., 50 (4):210-6 (2005)). Neither investigation, however, used a stringent analysis of suicide compared to mood disorder and suicide compared to controls to detect genes that might be most representative of suicide. This Example describes microarray gene expression profiles in the amygdala, anterior cingulate, and cerebellum in postmortem brains from BPD and MDD patients that committed suicide, focusing on mRNA expression levels of the molecules which regulate white-matter, oligodendrocyte, myelin, and other pathways. The genes identified here may be used as biomarkers for detecting and treating suicidal behavior.
[0258]Genes were discovered by selecting subjects run on U133P chips with mRNA quali...
example 2
Identification of Lithium Responsive Genes which are Dysregulated in BPD
[0261]This Example demonstrates that certain genes in non-human primates (healthy rhesus macaque monkey) are differentially expressed in response to treatment with the mood-stabilizing drug, lithium (Li), the drug of choice for the treatment of BP. Gene expression profiling was carried out on the anterior cingulate cortex (AnCg), dorsolateral prefrontal cortex (DLPFC), hippocampus (HC) and amygdala (AMY) of rhesus macaque monkeys, using the gene expression detection methods described herein, and compared to the human postmortem results described above. Table 2A shows the lithium-responsive genes which had been previously identified in the literature and which were confirmed by the present investigation. Table 2B shows genes that are newly identified as lithium-responsive in primates and which are also dysregulated in human subjects with bipolar disorder.
example 3
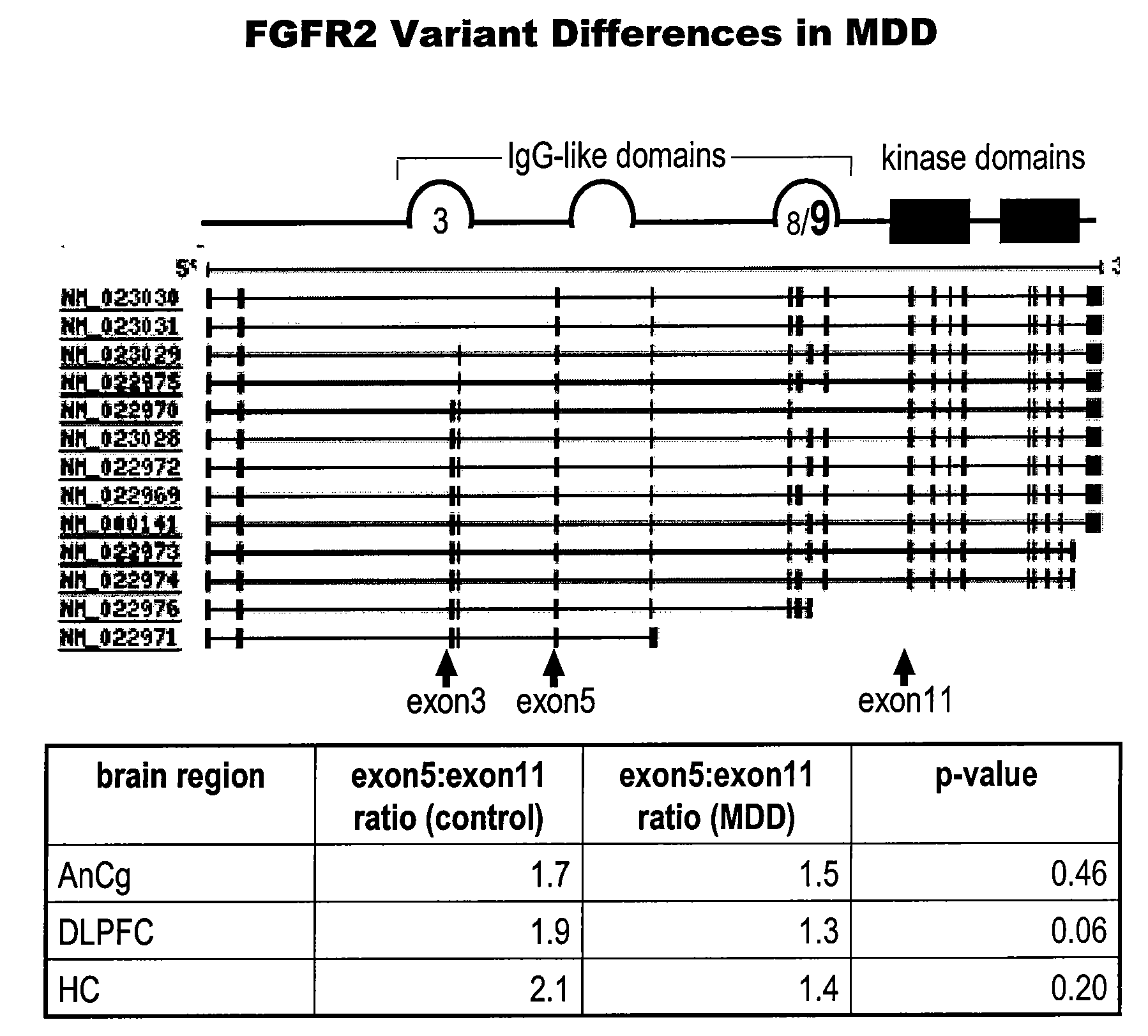
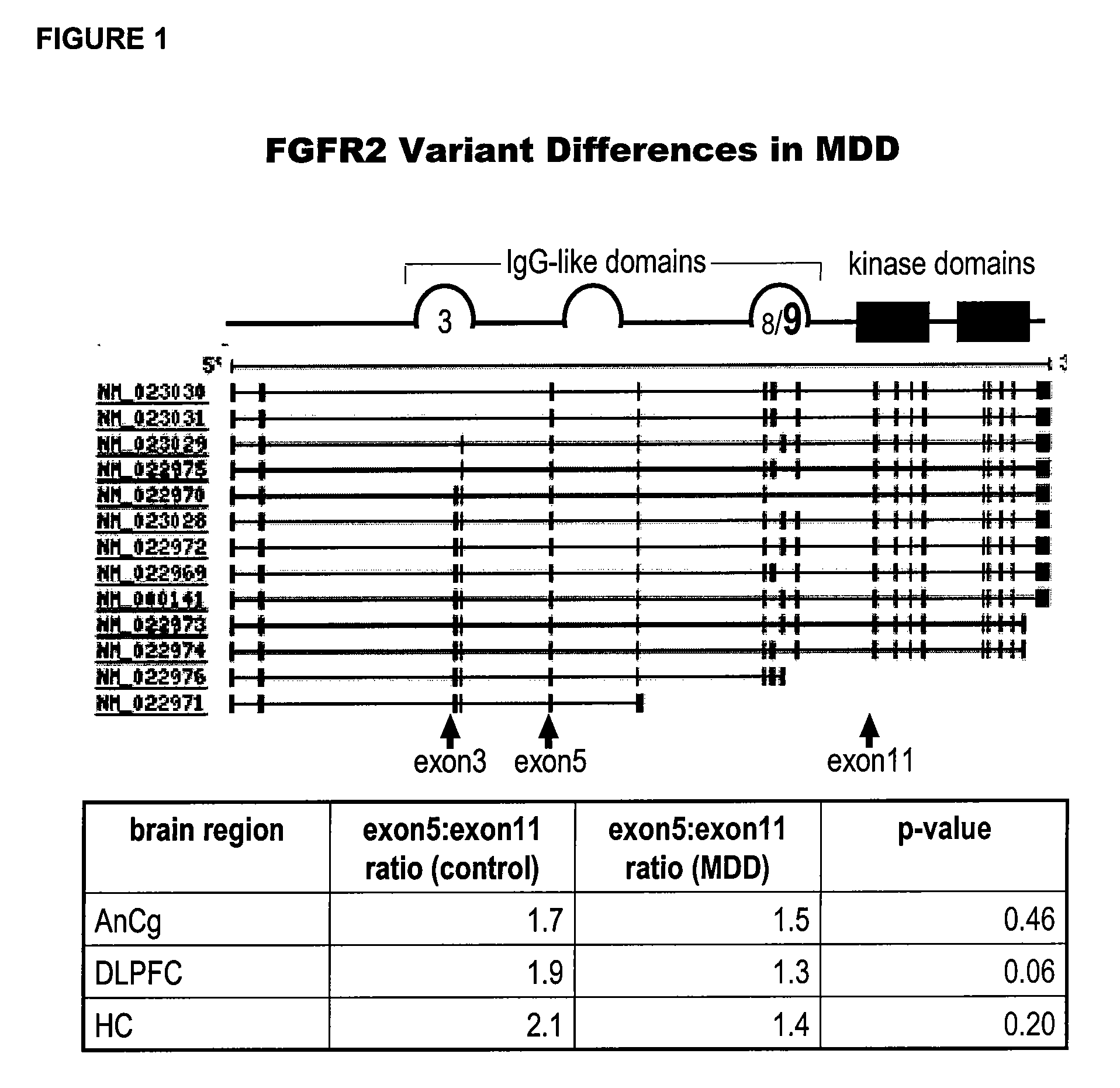
FGFR2 Variant Differences in Mood Disorders
[0262]The FGF receptor 2 (FGFR2) transcript is consistently found to be decreased in several brain areas of depressed subjects (see, e.g., U.S. patent application Ser. No. 10 / 701,263, filed Nov. 3, 2003, published as U.S. Pat. Publ. No. 20040152111-A1 on Aug. 5, 2004). The human FGFR2 gene contains 19 exons and produces as many as 13 splice variants. These variants fall into three main functional classes: first, variants that lack the transmembrane and tyrosine kinase domain which are thought to be soluble receptors; second, variants that contain the Ig IIIc type domain encoded by exon 9; and third, variants that contain the Ig IIIb type domain encoded by exon 8. The Ig III type domain confers ligand specificity and thus these latter two variants have different pharmacological profiles based on their use of the IIIc or IIIb domain. This Example describes PCR-based measurements of exons present in total RNA derived from human cortex (dorsola...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com