Nucleic acid amplification method
a technology of nucleic acid and amplification method, which is applied in the direction of microbiological testing/measurement, biochemistry apparatus and processes, fermentation, etc., can solve the problems of difficult primer design, complicated temperature control, and increased time loss in proportion to the number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
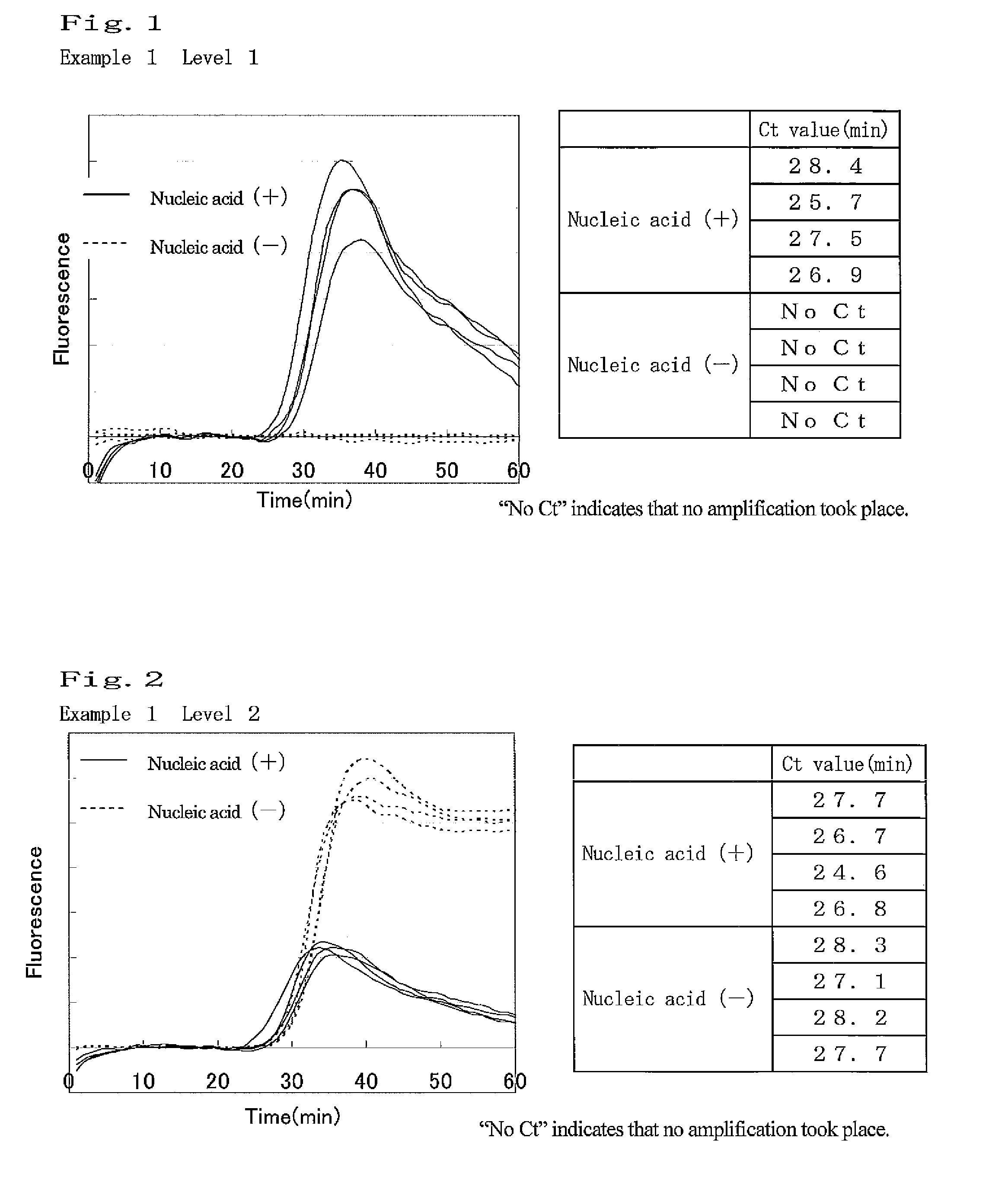
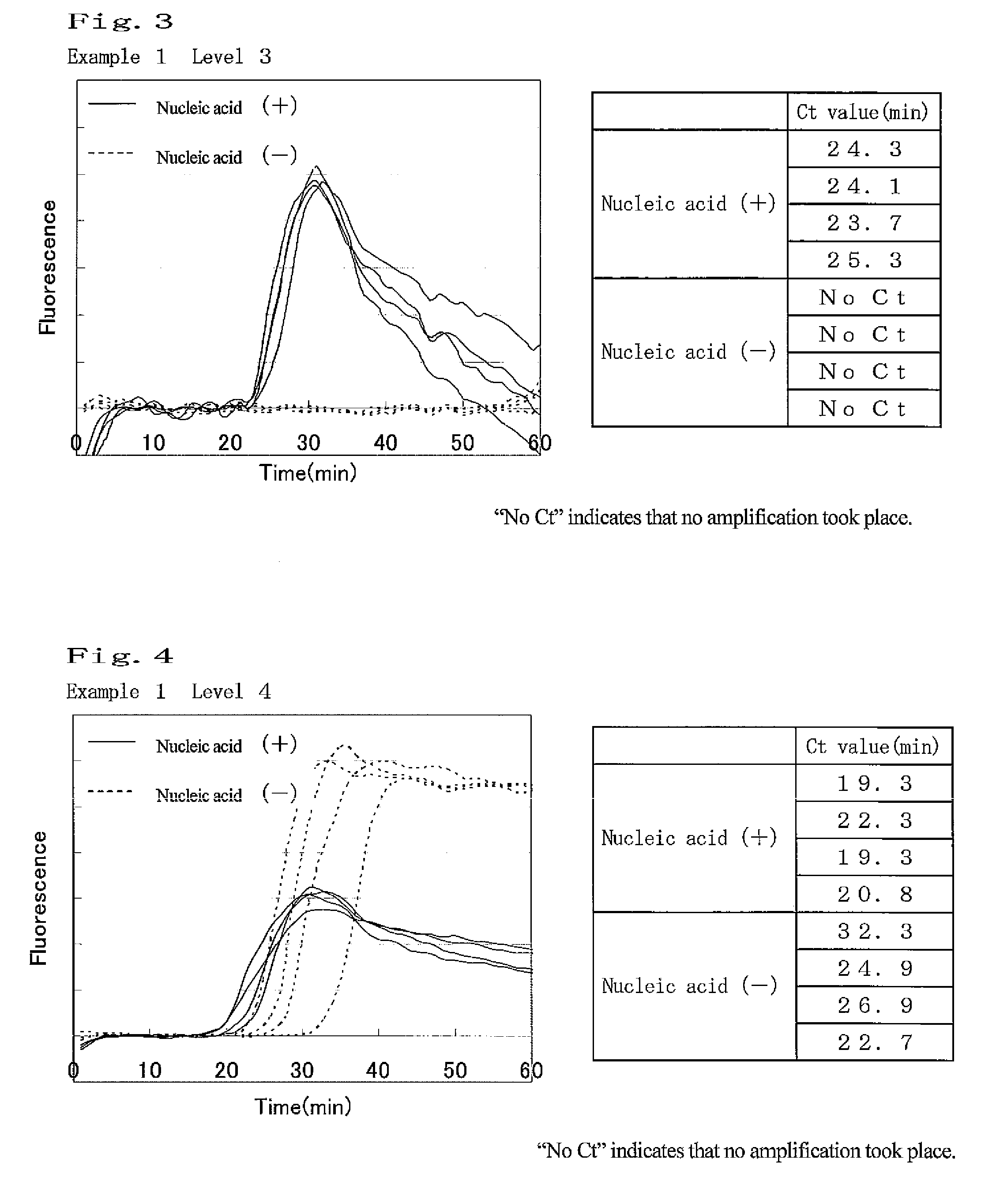
example 1
Amplification of Target Nucleic Acid Sequence in Human Gene
(1) Preparation of Nucleic Acid Specimen Solution Containing Target Nucleic Acid Fragment
[0115]7.5 ng of Human Genomic DNA (produced by Clontech) was heated at 98° C. for 3 minutes, and then a specific sequence in the target nucleic acid was amplified under the following conditions. As a negative control, a sample was also prepared by heating water under the same conditions.
[0116]The following 4 types of primers were designed, and purchased from Operon Biotechnologies. Each primer sequence is as shown below. Primers (1) and (2) are sequences of β-actin, and Primers (3) and (4) are complementary to sequences of β 2AR gene.
Primer (1):5′-GGGCATGGGTCAGAAGGATT-3′(SEQ ID NO: 1)Primer (2):5′-CCTCGTCGCCCACATAG-3′(SEQ ID NO: 2)Primer (3):5′-CTTGCTGGCACCCAATA-3′(SEQ ID NO: 3)Primer(4):5′-CCGGCGCATGGCTT-3′(SEQ ID NO: 4)
[0117]Tween 20 (Wako Pure Chemical Industries, Ltd.) was used as a surfactant. Tween 20 is polyoxyethylene(20) sorbita...
example 2
Effect of Concentration of the Surfactant
(1) Preparation of Nucleic Acid Specimen Solution Containing Target Nucleic Acid Fragment
[0139]7.5 ng of HumanGenomic DNA (produced by Clontech) was heated at 98° C. for 3 minutes, and then a specific sequence in the target nucleic acid was amplified under the following conditions. As a negative control, a sample was also prepared by heating water under the same conditions.
[0140]Primers (1) and (2) used in Example 1 were used as the primer.
Primer (1):5′-GGGCATGGGTCAGAAGGATT-3′(SEQ ID NO: 1)Primer (2):5′-CCTCGTCGCCCACATAG-3′(SEQ ID NO: 2)
[0141]Tween 20 (Wako Pure Chemical Industries, Ltd.) was used as a surfactant.
(2) Nucleic Acid Amplification Reaction
[0142]The amplification reaction was performed with the composition of a reaction solution shown below at 60° C. for 60 minutes. Bst. DNA polymerase (NEB (New England Biolabs)) was used as an enzyme.
[0143]
10 × Bst Buffer (Detergent Free)2.5 μL100 mM MgSO41.5 μL0% (v / v)-10% (v / v) Tween 201.25 μL ...
example 3
Effect of Type of the Surfactant
(1) Preparation of Nucleic Acid Specimen Solution Containing Target Nucleic Acid Fragment
[0149]7.5 ng of HumanGenomic DNA (produced by Clontech) was heated at 98° C. for 3 minutes, and then a specific sequence in the target nucleic acid was amplified under the following conditions. As a negative control, a sample was also prepared by heating water under the same conditions.
[0150]Primers (1) and (2) used in Examples 1 and 2 were used as the primer.
Primer (1):5′-GGGCATGGGTCAGAAGGATT-3′(SEQ ID NO: 1)Primer (2):5′-CCTCGTCGCCCACATAG-3′(SEQ ID NO: 2)
[0151]The following 9 types of substances were used as a surfactant, and experiment was carried out.
Level 1: Tween 40
[0152]Tween 40 is polyoxyethylene(20) sorbitan monopalmitate, and is a polyoxyethylene sorbitan fatty acid ester-based non-ionic surfactant. More particularly, Tween 40 is polyoxyethylene sorbitan fatty acid monoester. Tween 40 has HLB of 15.6, and is represented by the following formula. Tween 40...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| carbon number | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com