Use of a cysteine protease of Plasmodium vivax
a cysteine protease and plasmodium vivax technology, applied in the field of vivapain4, can solve the problems of high hinderance in the comprehensive study of i>p. vivax /i>cysteine proteases, etc., and achieves high efficiency, high unique substrate preference, and severe morbidity and mortality.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
In Silico Identification of Cysteine Protease Gene
[0073]1.1: Sequencing of Novel Cysteine Proteases
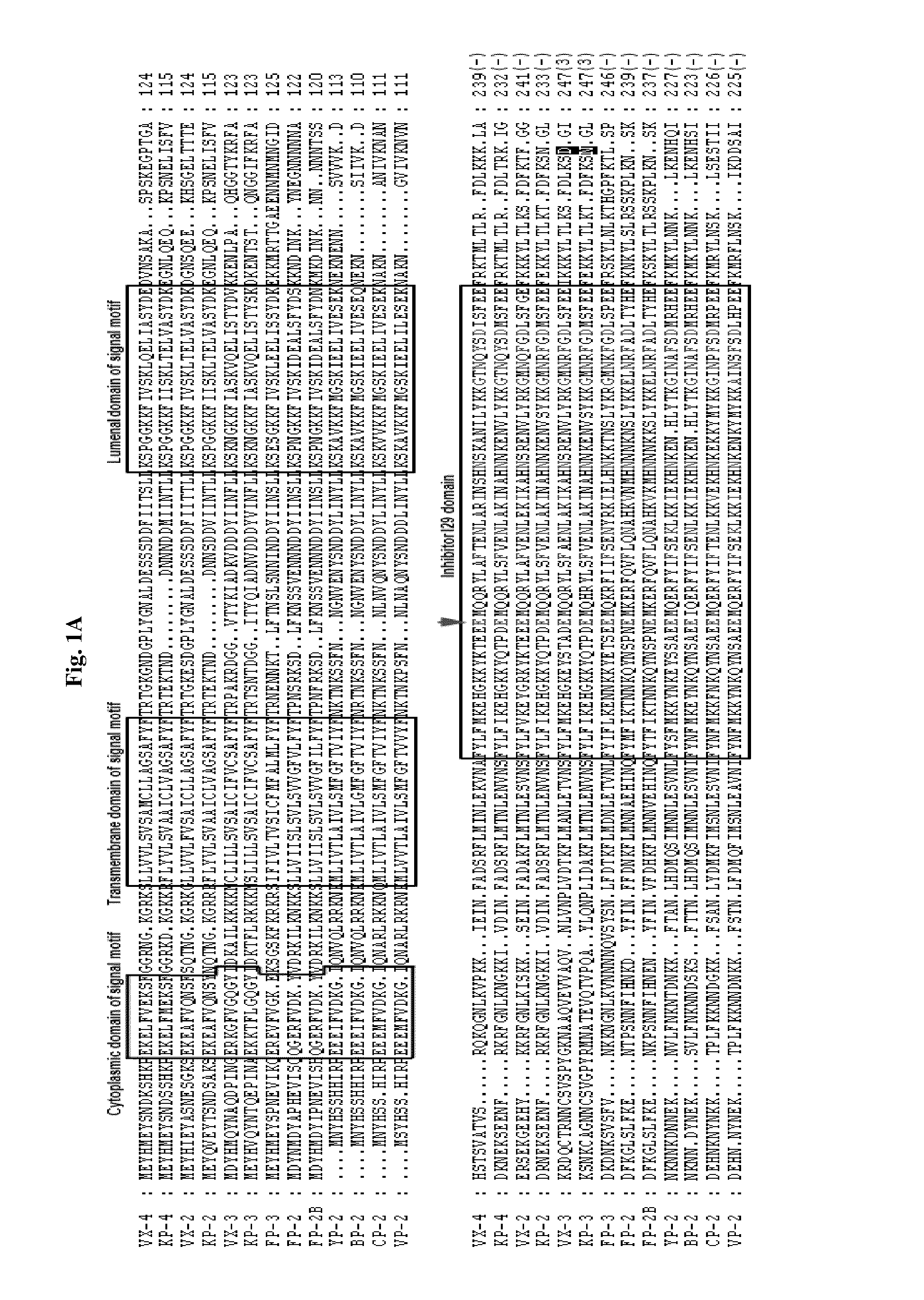
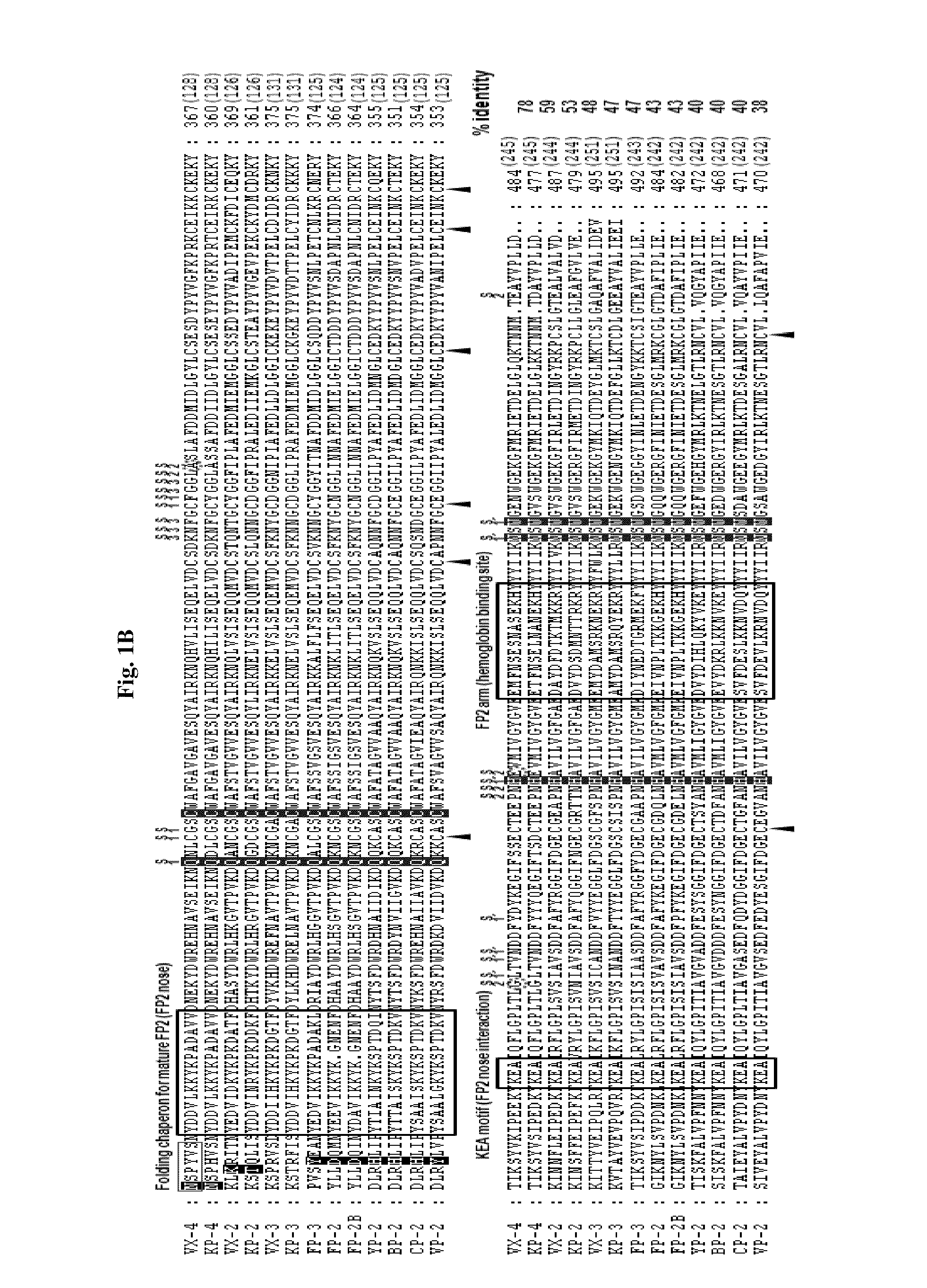
[0074]Genes putatively coding for cysteine proteases were identified from primate and rodent Plasmodium sequences deposited in PlasmoDB (http: / / plasmodb.org), TIGR (http: / / www.tigr.org), and GenBank (http: / / www.ncbi.nlm.nih.gov / ) through BLAST searches. The amino acid (AA) sequences of cysteine proteases of P. falciparum (FP-2 [XP—001347836], FP-2B [XP—001347832], and FP-3 [XP—001347833]), P. vivax (VX-2 [XP—001615274] and VX-3 [XP—001615273]), P. yoelii (yoelipain-2 [YP-2; XP—726900]), and P. berghei (bergheipain-2 [BP]-2 [XP—680416]), P. chabaudi (chabaupain-2 [CP-2], AAP43630), P. vinckei (vinckepain-2 [VP-2], AAL48319), P. knowlesi (knowlepain-2 [KP-2], CAQ39926; KP-3, CAQ39925; and KP-4, CAQ39924), P. falciparum (FP-2, XP—001347836; FP-2B, XP—001347832; and FP-3, XP—001347833), and P. vivax (VX-2 [XP—001615274] and VX-3 [XP—001615273]) were used in multiple queries, with a thresho...
example 2
Expression and Refolding of a Recombinant VX-4 (rVX-4)
[0088]The open reading frame (ORF) of VX-4 was amplified with forward (5′-ATGGAATATCACATGGAGTACTCGAAC-3′, SEQ ID NO: 3) and reverse (5′-CTAGTCAAGCAGGGGGACGTACGCCTC-3′, SEQ ID NO: 4) primers using Ex Taq DNA polymerase (Takara, Japan) and P. vivax genomic DNA (100 ng) isolated from a Korean patient (a generous gift from Dr. J S Yeom [Sungkyunkwan University Hospital]). The PCR conditions were as follows:
[0089]94° C. 5 min
[0090]94° C. 1 min, 50° C. 1 min, 72° C. 2 min: 30 cycles
[0091]72° C. 10 min
[0092]The product was gel-purified, ligated into the pCR2.1 vector (Invitrogen, CA) nd transformed into competent E. coli Top10 cells (Invitrogen) by heat shock (ice 30 minutes, 42° C. 45 seconds, ice 5 minutes). The purification, ligation and transformation were conducted according to a manual provided by Invitrogen. The nucleotide sequence was determined with an ABI PRISM 377 DNA sequencer (Applied Biosystems, CA).
[0093]The obtained DNA ...
example 3
N-Terminal Amino Acid Sequencing
[0103]The fully processed rVX-4 was separated by 12% SDS-PAGE. The protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) and stained with Coomassie blue. The band was excised and subjected to protein sequencing on an ABI model 477A protein sequencer and an ABI model 120A PTH analyzer (Applied Biosystems) at the Korea Basic Science Institute (Daejeon, Korea). The obtained N-terminal amino acid sequence is shown in red box of FIG. 1B, from which mature rVX-4 is initiated.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com