Piggybac transposon variants and methods of use
a technology of piggybac and variants, applied in the field of piggybac transposon variants and methods of use, can solve the problems of method limitations, size constraints of dna condensing reagents and virus-mediated strategies, and the limited amount of nucleic acids that can be transfected into cells, so as to achieve a lower rate of integration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of Integration-Defective PiggyBac Variants
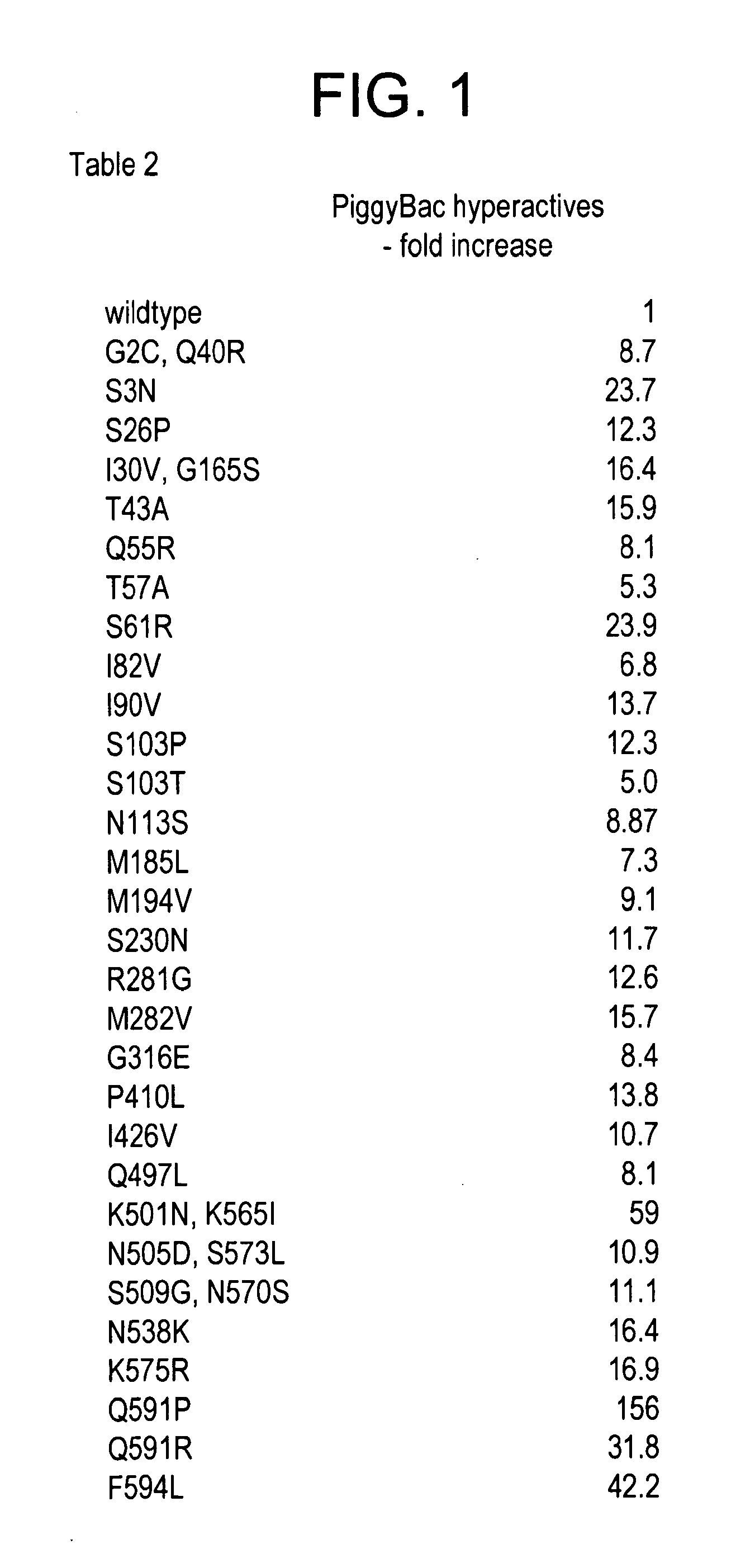
[0191]The present experiments describe screening and identification of excision-hyperactive piggyBac transposons using a version of the Cherry gene which produces a red fluorescent protein. A copy of the piggyBac transposon was put into the gene, inactivating it such as the cells are NOT red. However, piggyBac excision restores the gene, leading to the production of the red fluorescent protein. Accordingly, increased red colony color identifies mutants that excise better.
[0192]A large collection of mutant transposase genes was made using mutagenic PCR which was cloned into an expression vector in yeast. Colonies containing individual mutants were grow up on an agar plate and were then examined with red fluorescent light. FIG. 4 shows excision hyperactives that have been isolated to date.
[0193]In certain preferred embodiments, the integration defective piggyBac comprises an amino acid change in the wild type piggyBac sequence c...
example 2
Identification of Hyperactive Variants
[0195]Using the integration defective piggyBac mutants as a starting point, the present inventors have identified hyperactive piggyBac transposon mutants. The yeast excision assay that was developed as described in Mitra R. et al. (piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. Apr 9; 27(7):1097-109. Epub 2008 Mar. 20) was used to identify the hyperactive mutants. The piggyBac ORF was mutagenized by mutagenic PCR using primers flanking the ORF as the expression construct and then recovered transformants by co-transformation of the PCR product with a gapped piggyBac plasmid into the yeast assay strain containing a ura− to ura+ cassesette in which transposon excision results in formation of ura+ colonies. Following recovery of transformants on SC-Trp-His plates, colonies were resuspended in water and spotted onto plates lacking uracil to identify excisions. By comparison to the number of ura+ colonies from the mutage...
example 3
Induced Pluripotent Stem Cell Generation Using the Hyperactive Transposon
[0198]In certain exemplary embodiments, the hyperactive piggyBac transposons can be used to created induced pluripotent stem cells using a minimal set of genes. In particular, Oct 3 / 4, Sox2, Klf4 and c-myc are used as a minimal set of genes. Takahashi et al. (Cell, 131, 861-872, Nov. 30, 2007), incorporated by reference in its entirety herein, teach methods of generating induced pluripotent stem cells (iPS) from human dermal fibroblasts using Oct 3 / 4, Sox2, klf4, and c-Myc.
Other Embodiments
[0199]From the foregoing description, it will be apparent that variations and modifications may be made to the invention described herein to adopt it to various usages and conditions. Such embodiments are also within the scope of the following claims.
[0200]The recitation of a listing of elements in any definition of a variable herein includes definitions of that variable as any single element or combination (or subcombination...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Nucleic acid sequence | aaaaa | aaaaa |
| Level | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com