Cell response assay for cancer and methods of producing and using same
a cell response and cancer technology, applied in the field of cell response assays, can solve the problems of increasing the difficulty of measurement, reducing the fluorescence signal of nanoparticles, and poor sensitivity of traditional fluorescent labels for detecting rare cell analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Multiplexed Assay for Breast Cancer
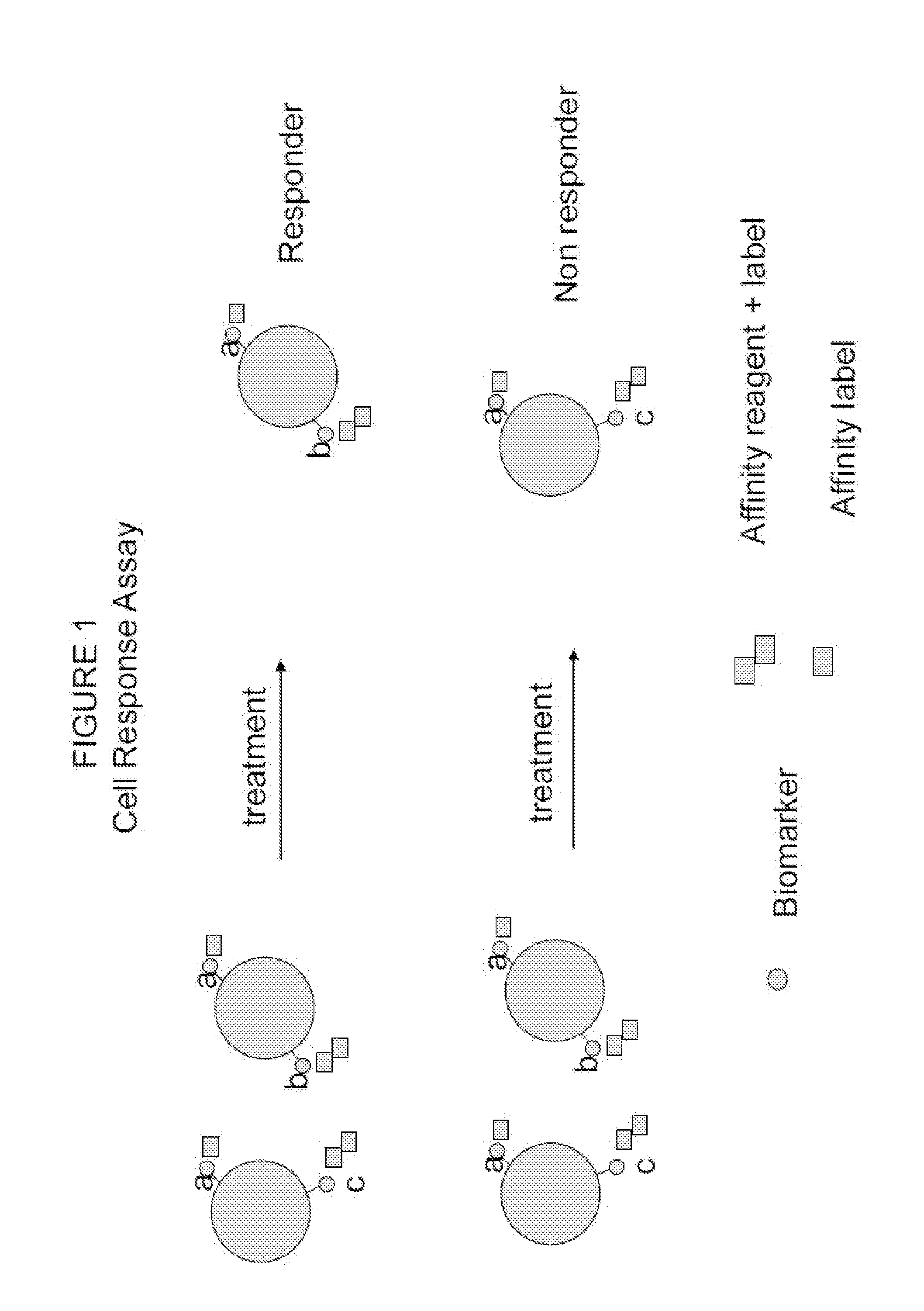
[0079]The following procedure shows the multiplexing of Biomarkers A (cancer cell type), B (metastatic potential) and C (chemo resistance) using a unique combination of affinity reagents with fluorescent labels and an affinity label to provide a multiplexed assay for breast cancer. This allows the simultaneous measurement of cancer cell type, chemo resistance nature, and metastatic potential, as described herein above, in combination with a marker indicating the presence of cell nucleus (utilizing a nucleic acid binding probe). The data demonstrated that four simultaneous signals can be measured with this combination but not with the traditional combinations of the prior art. The presently disclosed and claimed inventive concept(s) has the potential to measure up to eight signals utilizing various fluorophores taught herein.
[0080]The procedure to test the method was to collect 4 mL normal whole blood fresh into a VACUTAINER® tube (BD 6 mL VACUTAINE...
example 2
Cell Response Assay for Breast Cancer
[0085]A novel cell response assay for breast cancer was developed by using HER2 / neu as a tissue type marker for carcinoma breast cancer (Biomarker A) that is always present in the disease state. Biomarker B for metastatic potential utilized uPA and PAI antigen; these are markers of tumor invasiveness and are present in aggressive cancer cell differentiation and growth and increase with metastatic invasiveness potential of the cells. Biomarker C for chemo resistance utilized PL2L piwi like antigen (see for example, Gao et al., 2008).
[0086]The procedure shown in Example 1 was used to measure cancer cells before and after treatment with and without camptothecin. Camptothecin is known to activate apoptosis in cancer cells and kill cancer cells like a chemotherapy agent (See Gupta, 1997). Cells were tested by the cell response assay and by a traditional prior art assay. The novel “cell response assay” utilized HER2 / neu as Biomarker A for cell type, th...
example 3
Multiplexed Assay for Prostate Cancer
[0089]The following procedure shows the multiplexing of Biomarkers A (cancer cell type), B (metastatic potential) and C (chemo resistance) using a unique combination of affinity reagents with fluorescent labels and an affinity label to provide a multiplexed assay for prostate cancer. This allows the simultaneous measurement of cancer cell type, chemo resistance nature, and metastatic potential, as described herein above, in combination with a marker indicating the presence of cell nucleus (utilizing a nucleic acid binding probe). The data demonstrated that four simultaneous signals can be measured with this combination but not with the traditional combinations of the prior art. The presently disclosed and claimed inventive concept(s) has the potential to measure up to eight signals utilizing various fluorophores taught herein.
[0090]The procedure to test the method was to collect 4 mL normal whole blood fresh into a VACUTAINER® tube (BD 6 mL VACUT...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical resistance | aaaaa | aaaaa |
| Wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com