Diagnostic method
a cancer and prognosis technology, applied in the field of cancer diagnosis, prognosis or monitoring, can solve the problems of methods plagued by the number of false positives generated, and achieve the effect of assessing the effectiveness of treatment in the individual and predicting the susceptibility of individuals to cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
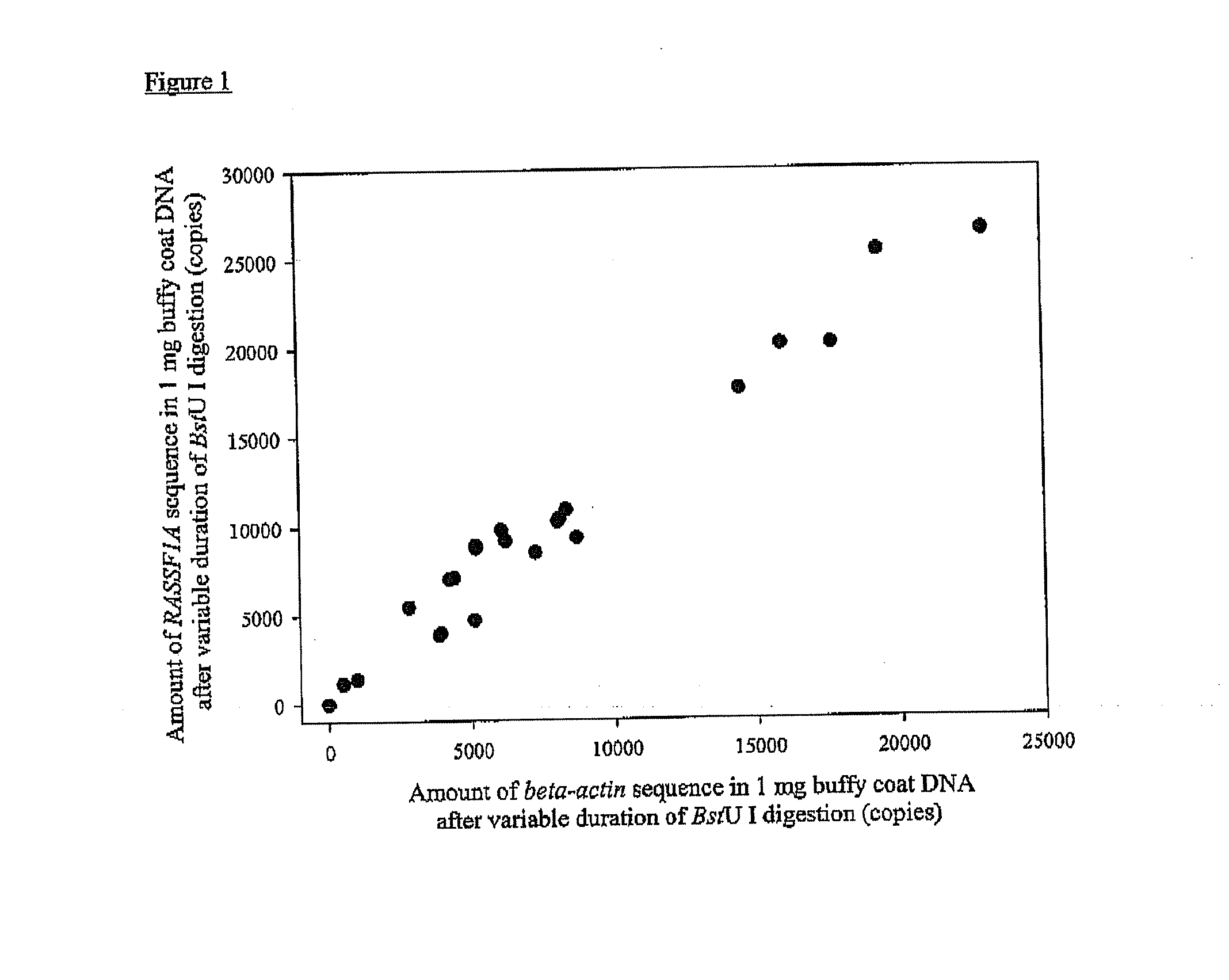
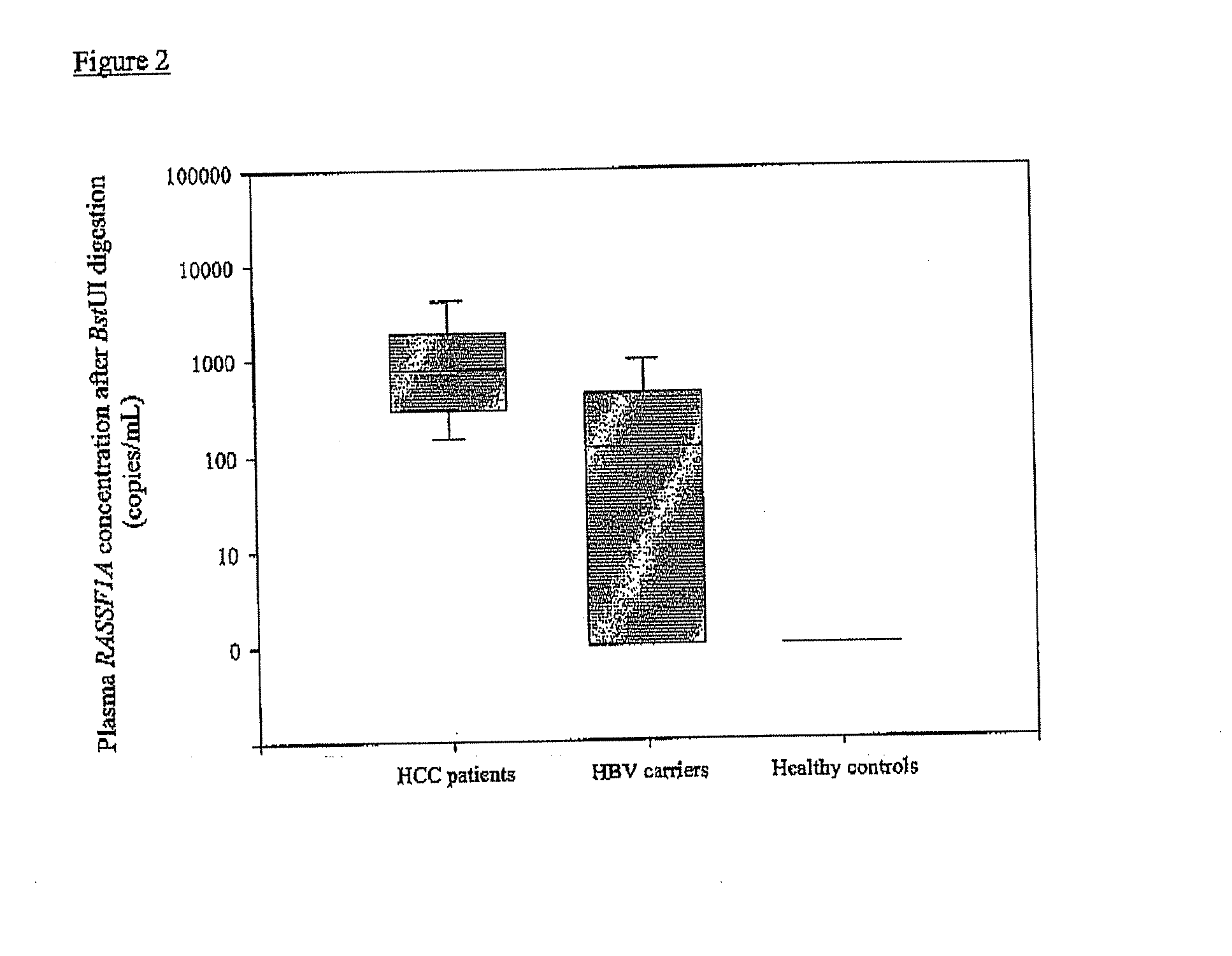
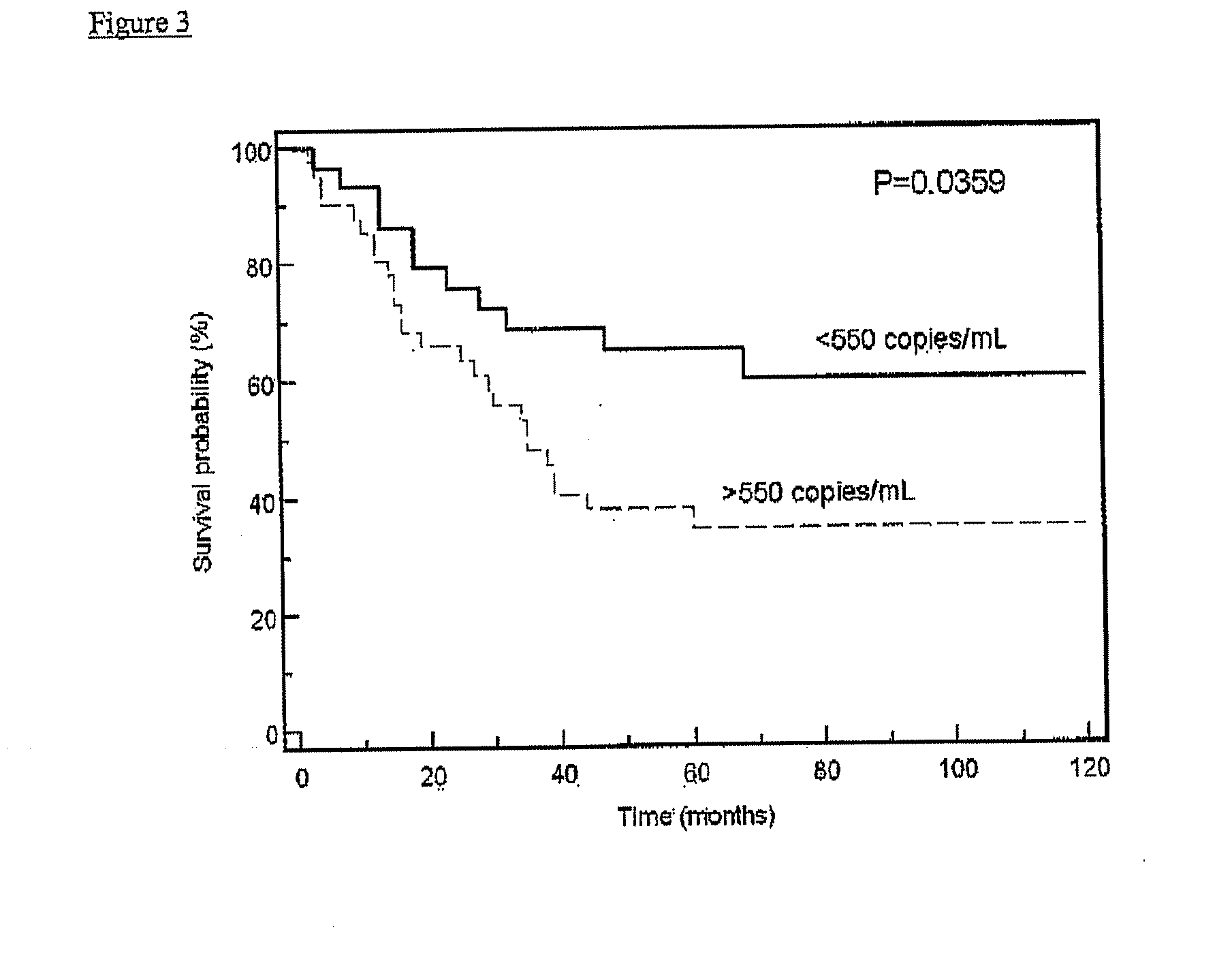
[0156]Sixty-three hepatocellular carcinoma (HCC) patients, as well as two groups of controls, were recruited. The first control group consisted of 15 healthy control subjects without chronic hepatitis B infection. The second control group consisted of chronic hepatitis B carriers. For each HCC patient, one sex-and age-matched chronic hepatitis B carrier was recruited as a control. Chronic hepatitis B carriers had increased risk of developing HCC and, thus, would be the target group for HCC screening. Four milliliters of venous blood were collected from each study subject into EDTA-containing tubes. Blood samples were centrifuged at 1,600 g for 10 min and the supernatant was re-centrifuged at 16,000 g for 10 min. DNA was then extracted from 800 μL of plasma using the QIAamp mini kit (Qiagen, Hilden, Germany) and eluted with 50 μL of H2O. Thirty-five microliters of plasma DNA were digested with 100 U of BstU I enzyme, in 1× digestion buffer at 60° C. for 16 hours.
[0157]The concentrati...
PUM
| Property | Measurement | Unit |
|---|---|---|
| incubation time | aaaaa | aaaaa |
| β | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com