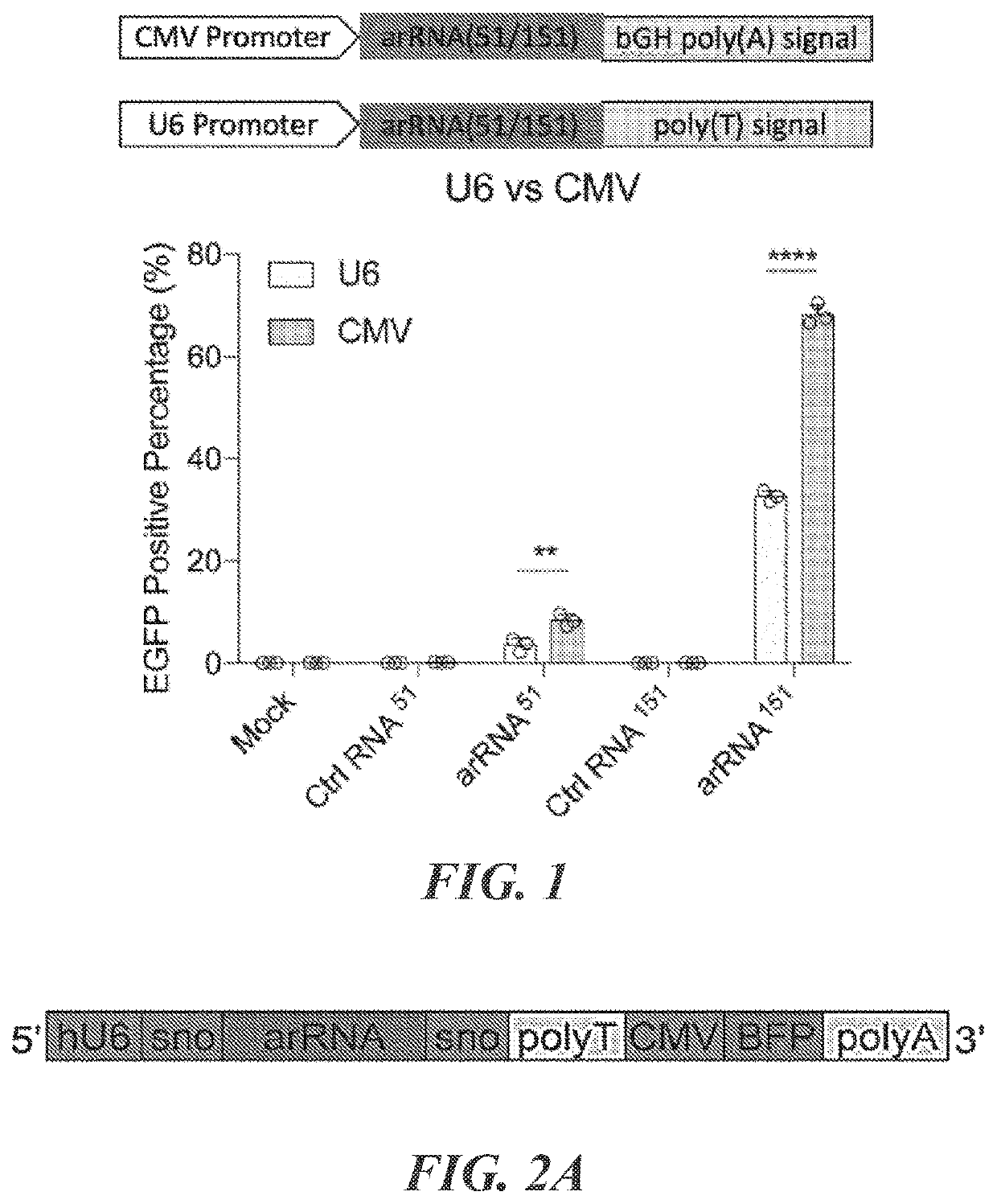
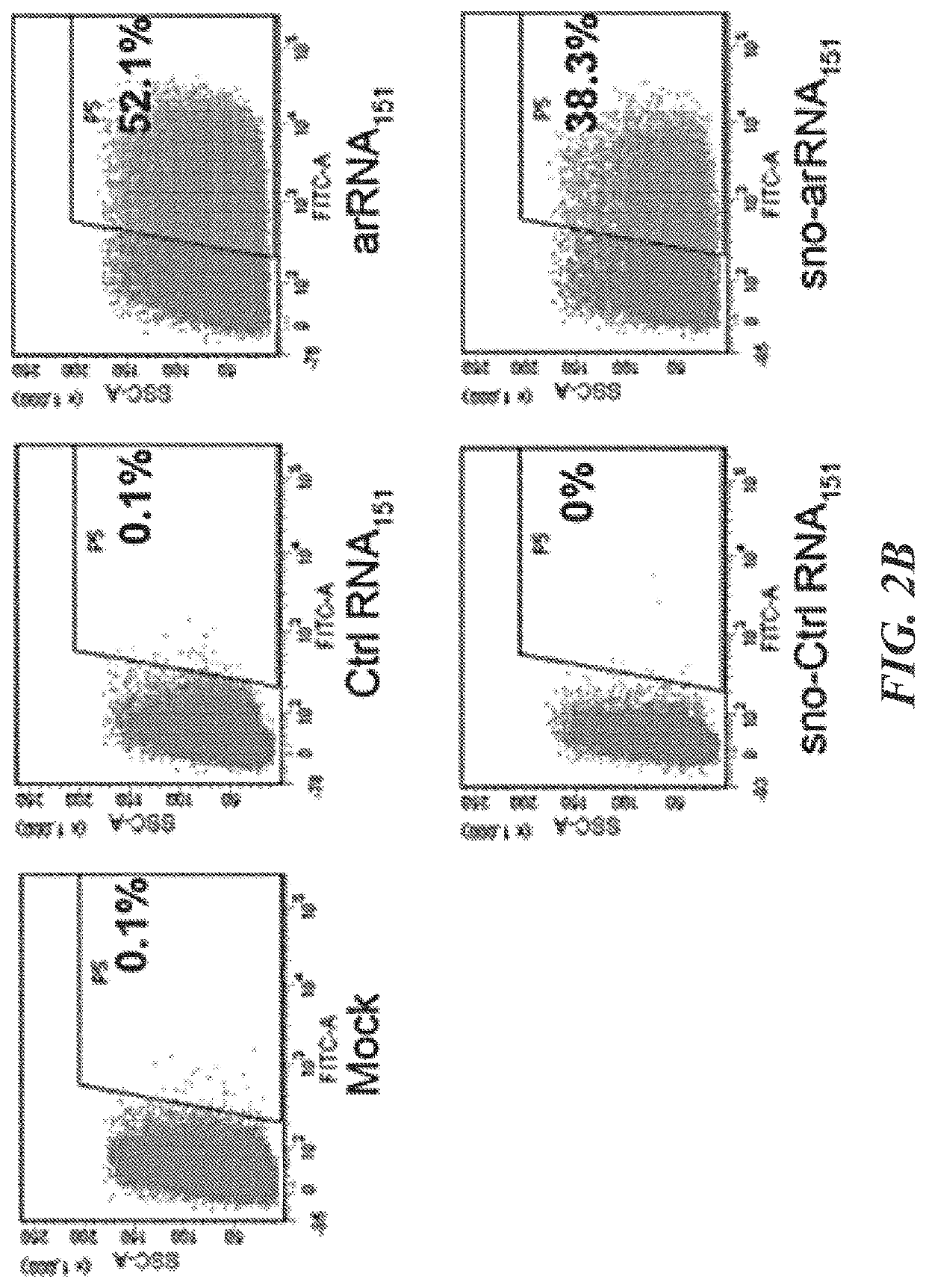
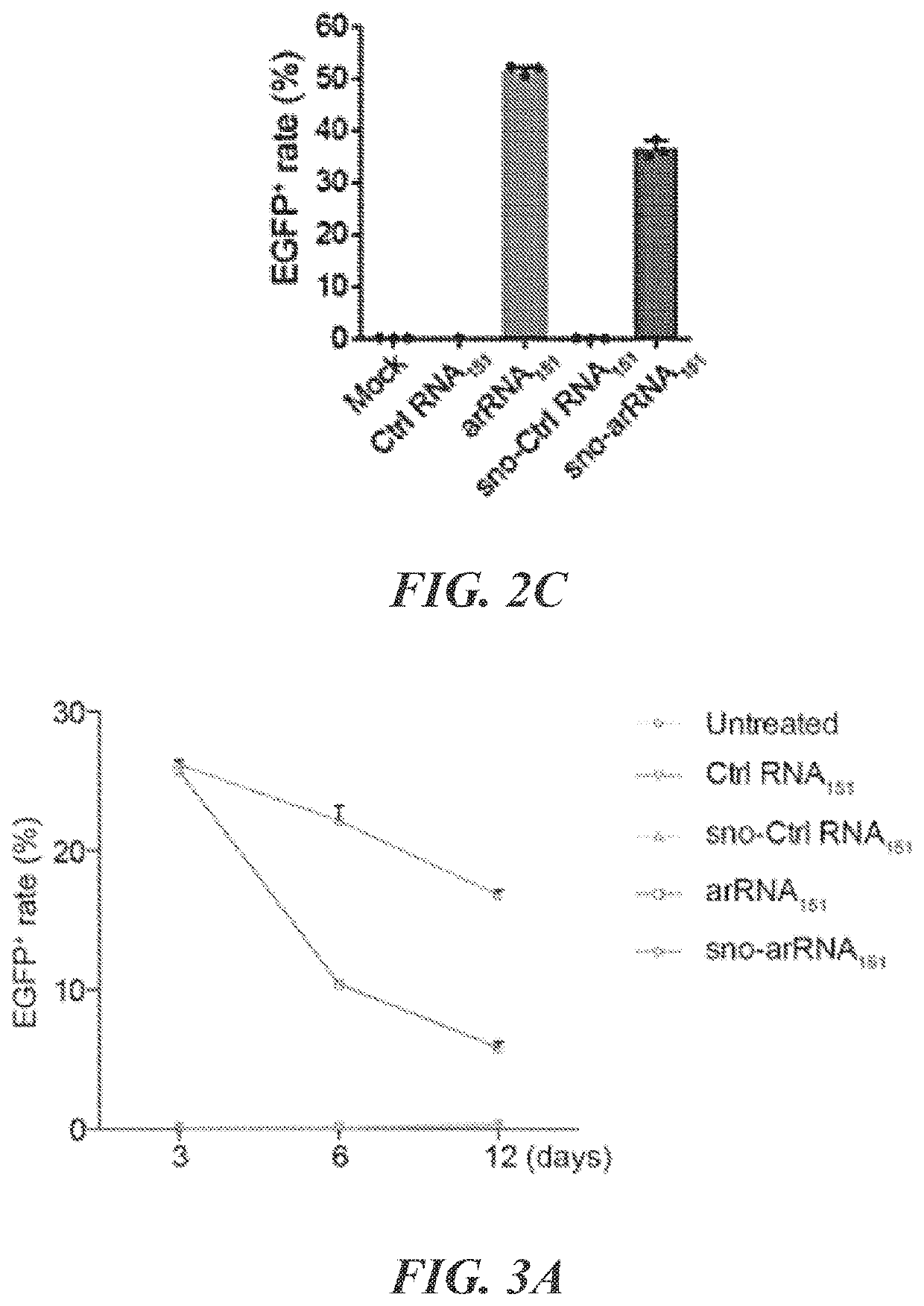
Targeted RNA editing by leveraging endogenous adar using engineered rnas
a technology of rna editing and endogenous adar, which is applied in the direction of dsdna viruses, viruses/bacteriophages, genetic material ingredients, etc., can solve the problems of ectopic expression of proteins or their domains of non-human origin that have potential risk of eliciting immunogenicity, and the current adar-mediated rna editing technology has certain limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
embodiment 1
2. The method of embodiment 1, wherein the dRNA further comprises a 3′ ligation sequence and a 5′ ligation sequence.
embodiment 2
3. The method of embodiment 2, wherein the 3′ ligation sequence and the 5′ ligation sequence are at least partially complementary to each other.
4. The method of embodiment 2 or embodiment 3, wherein the 3′ ligation sequence and the 5′ ligation sequence are about 20 to about 75 nucleotides in length.
5. The method of any one of embodiments 1-4, wherein the dRNA is circularized by RNA ligase RtcB.
embodiment 4
6. The method of embodiment 4, wherein the RNA ligase RtcB is expressed endogenously in the host cell.
7. The method of any one of embodiments 1-6, wherein the dRNA is a circular RNA.
8. The method of any one of embodiments 1-6, wherein the dRNA is a linear RNA capable of forming a circular RNA.
9. The method of any one of embodiments 1-6, wherein the method comprises introducing a construct comprising a nucleic acid encoding the dRNA into the host cell.
10. The method of claim 9, wherein the construct further comprises a 3′ twister ribozyme sequence linked to the 3′ end of the nucleic acid encoding the dRNA and a 5′ twister ribozyme sequence linked to the 5′ end of the nucleic acid encoding the dRNA.
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com