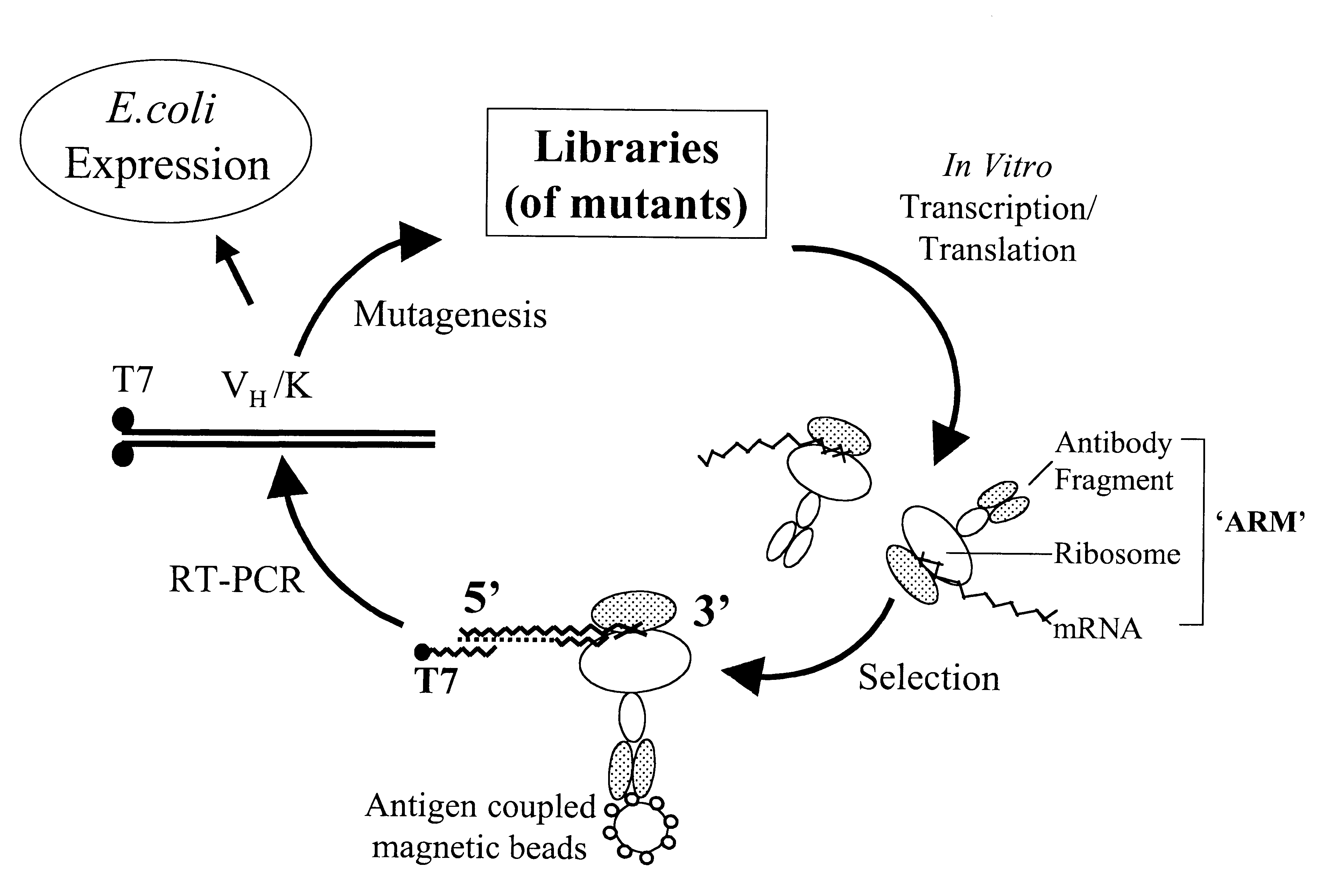
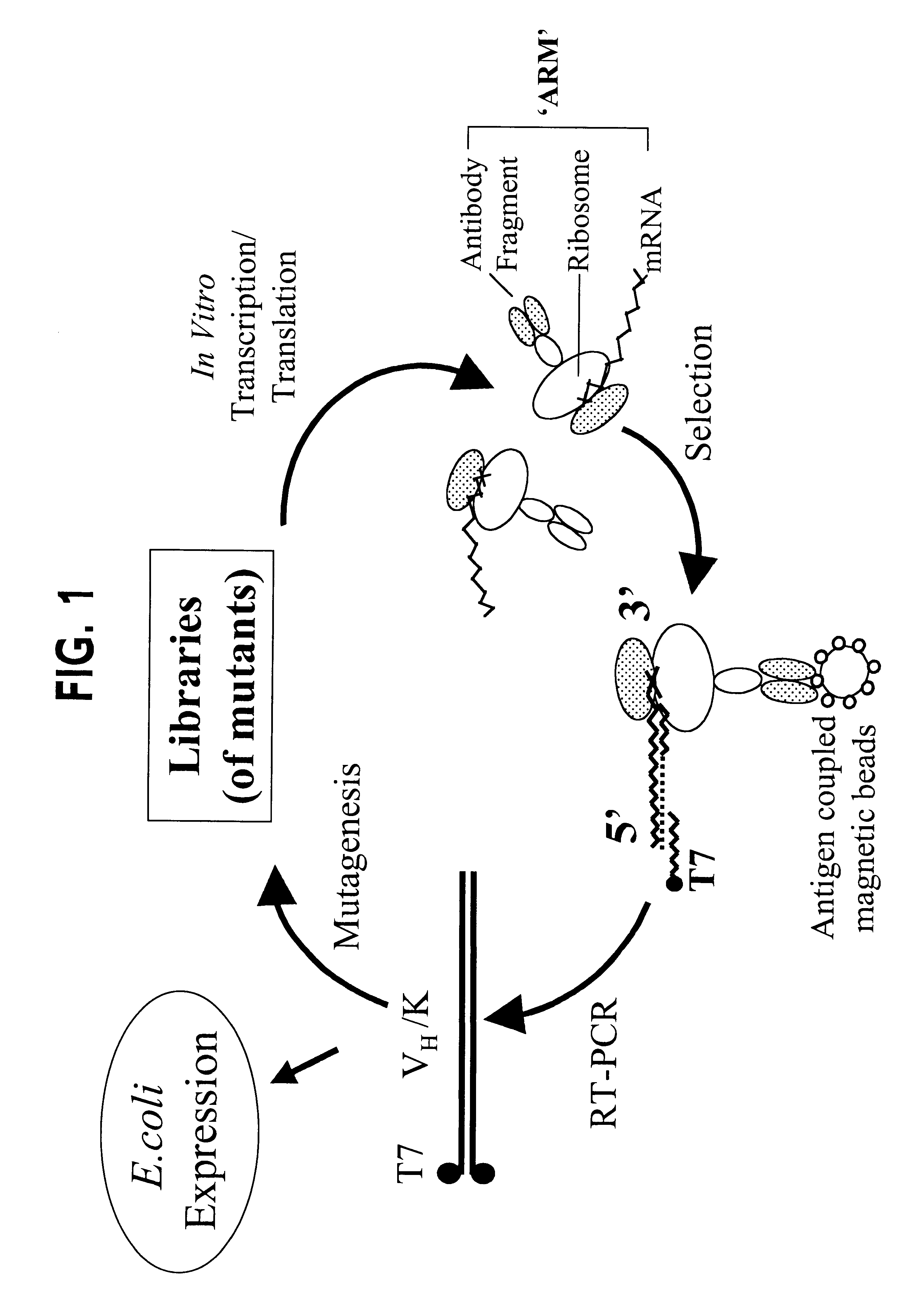
Ribosome complexes as selection particles for in vitro display and evolution of proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Recovery of DNA by RT-PCR on the Ribosome Complex and Use of 2- or 3-Primer Combinations
In the ARM method (FIG. 1), the ribosome is stalled and the stable complex (nascent protein-ribosome-mRNA) forms because of the absence of a stop codon at the 3' end of the message. Since the ribosome is stalled at the 3' end of the mRNA, the latter should be inaccessible to a 3' primer and / or to reverse transcriptase, necessitating the use of an upstream primer in the recovery of cDNA. This is confirmed by the experiment in FIG. 3. When full length DB3 DNA, lacking the 3' stop codon, was transcribed and the mRNA translated in vitro and selected with progesterone-BSA beads, cDNA recovery showed that the 3' end of the mRNA was not available for priming in RT-PCR, whereas an upstream primer (D2, FIG. 2A) successfully recovered the cDNA. Likewise, in a second cycle, D2 was no longer effective and a primer further upstream (D3, FIG. 2A) was required. Thus, the concept of a ribosome bound to the 3' en...
example 2
Antigen-Specific ARM Selection
To demonstrate antigen-specific ARM selection, DB3.sup.R V.sub.H / K was translated in vitro and ARMs exposed to magnetic beads coupled either to progesterone-11.alpha.-BSA, testosterone-3-BSA or BSA alone. After RT-PCR, a single DNA fragment was detected only from progesterone-11.alpha.-BSA coupled beads (FIG. 5A, tracks 2,4,6), consistent with the known specificity of DB3.sup.R V.sub.H / K. The recovered fragment was further confirmed as DB3.sup.R by sequencing. No bands were obtained when PCR alone, rather than RT-PCR, was carried out on the progesterone-11.alpha.-BSA beads after selection (FIG. 5A, tracks 3,5,7), or when the procedure was performed with nontranslated DB3.sup.R mRNA (FIG. 5A, track 1). Thus, the band recovered by RT-PCR is derived from mRNA selected via the functional antibody combining site of DB3.sup.R and not from DNA contamination or mRNA carryover.
example 3
Inhibition by Free Antigen of ARM Binding to Immobilised Antigen Demonstrates Correct Folding of the VH / K on the Ribosome
Inhibition by free steroids can be used to demonstrate the correct folding and functional activity of the ARM complex (FIG. 6). The inhibition of DB3 V.sub.H / K expressed as an ARM, using different steroidal inhibitors, is indistinguishable from that of native DB3 and recombinant V.sub.H / K. Furthermore, the 50% inhibition by progesterone-11.alpha.-HMS at 1 ng (2.5 nM) indicates an affinity very close to that of DB3 (data not shown).
The free steroid inibitors were added to the DB3 ARM mixture in order to block binding to the progesterone-coated beads. They are progesterone-11.alpha.-hemisuccinate (HMS) (P11), progesterone-3-carboxymethyloxime (P3); progesterone-6-HMS (P6) and progesterone-21-HMS (P21). The inhibition of free DB3 V.sub.H / K in an ELISA reaction is shown on the right, with the efficiency of the steroids in the order P11>P3>P6>P21. A very similar ord...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com