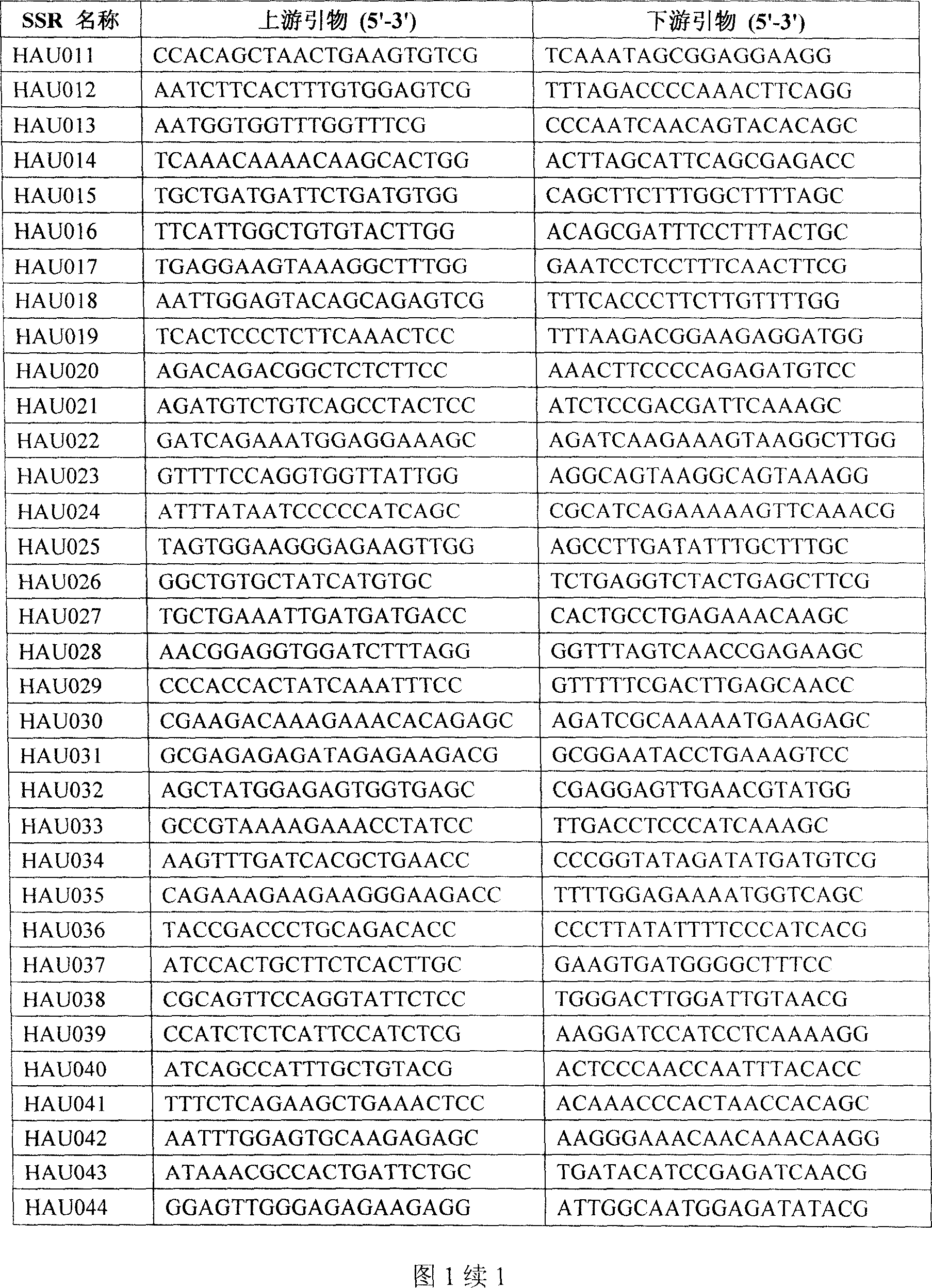
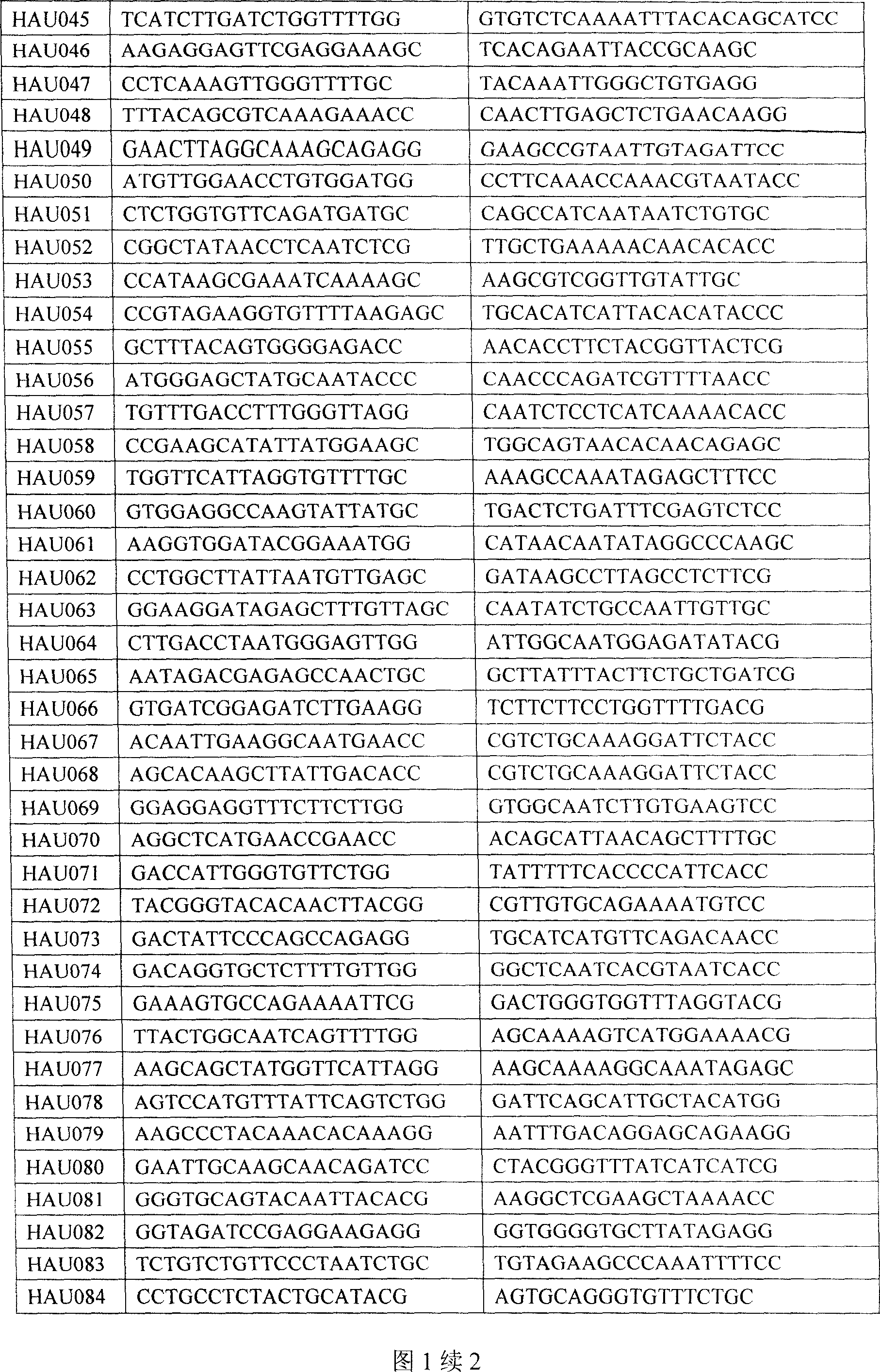
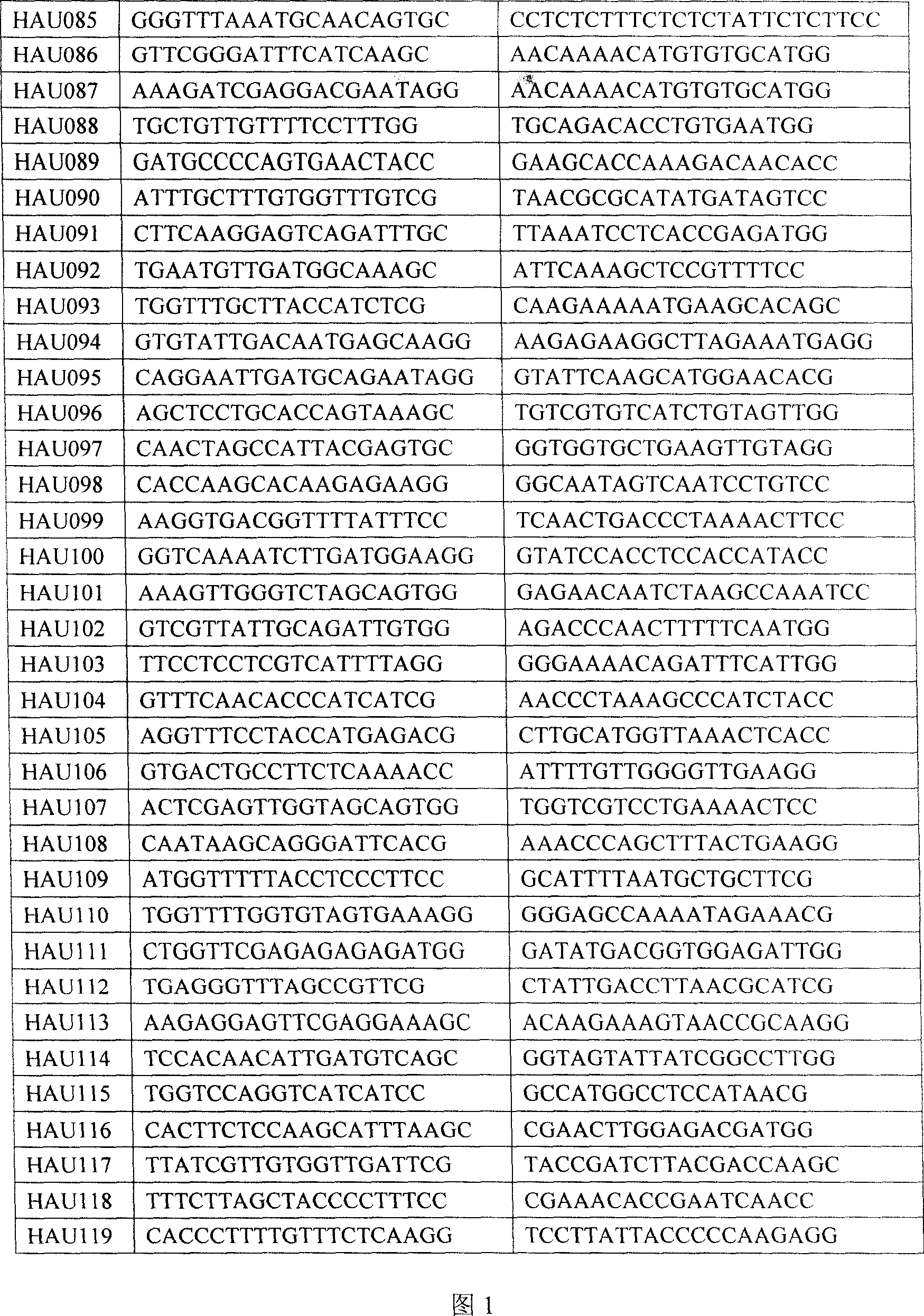
Prepn process and application of sea island cotton EST SSR marker
A sea-island and marker technology, applied in the preparation of sea-island cotton EST-SSR primer sequences and QTL mapping, can solve the problem of small data volume of African cotton and sea-island cotton
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1, get 36 parts of cotton seed materials (as shown in Figure 2), its genotype is AA13, DD11 AAD12) as the material of genetic diversity analysis, and described material is planted in the field;
[0041] 2. Get the tender green leaves from the field material plant and extract its total DNA. The specific method is with reference to Paterson et al. in 1994 (Paterson A H, Curt L B, Wendel J F, A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP and PCR analysis. Plant Mol Bil Rep, 1993, 11: 112-127) reported method extraction;
[0042] 3. Utilize the sea island cotton EST-SSR primer sequence among the present invention to analyze, and concrete steps are as follows:
[0043] 3.1 HAU primer marker analysis:
[0044] PCR amplification reaction system is 20μL: 25ng DNA, 4μmol upstream primer, 4μmol downstream primer, 1×buffer (10mM Tris-Hcl, 50mM Kcl, pH8.3), 2mmol Mg2 + , 0.25mmol dNTPs, and 0.8 U Taq polymerase.
[0045] The PCR amplifica...
Embodiment 2
[0058] 1. Parent Emian 22, Pima3-79 (provided by the Cotton Research Institute of the Chinese Academy of Agricultural Sciences) and BC 1 The population is used as the starting material for the construction of the genetic linkage map, and the material is planted in the field;
[0059] 2. Extract its total DNA from the tender green leaves on the field material plant;
[0060] 3. Utilize the sea island cotton EST-SSR primer sequence among the present invention to analyze, and concrete steps are as follows:
[0061] 3.1 HAU primer marker analysis:
[0062] The PCR amplification reaction system was 20 μL: 25ng DNA, 4 μmol upstream primers, 4 μmol downstream primers, 1 × buffer (10mM Tris-Hcl, 50mM Kcl, pH8.3), 2mmol Mg2+, 0.25mmol dNTPs, and 0.8 U Taqpolymerase.
[0063] The PCR amplification reaction program was as follows: denaturation at 94°C for 3 minutes, followed by 34 cycles (denaturation at 94°C for 50 seconds, annealing at 57°C for 45 seconds, extension at 72°C for 1 minut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com