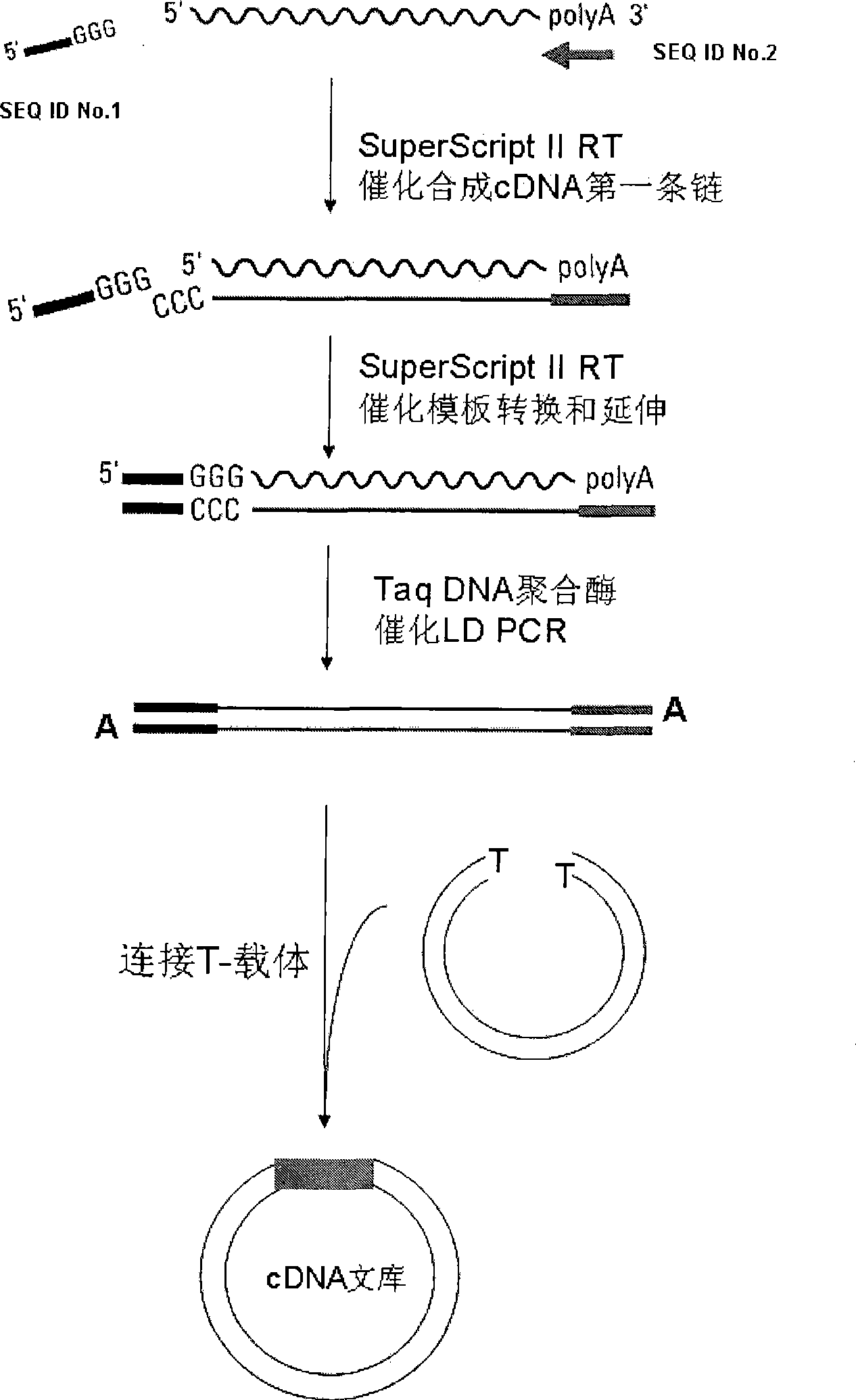
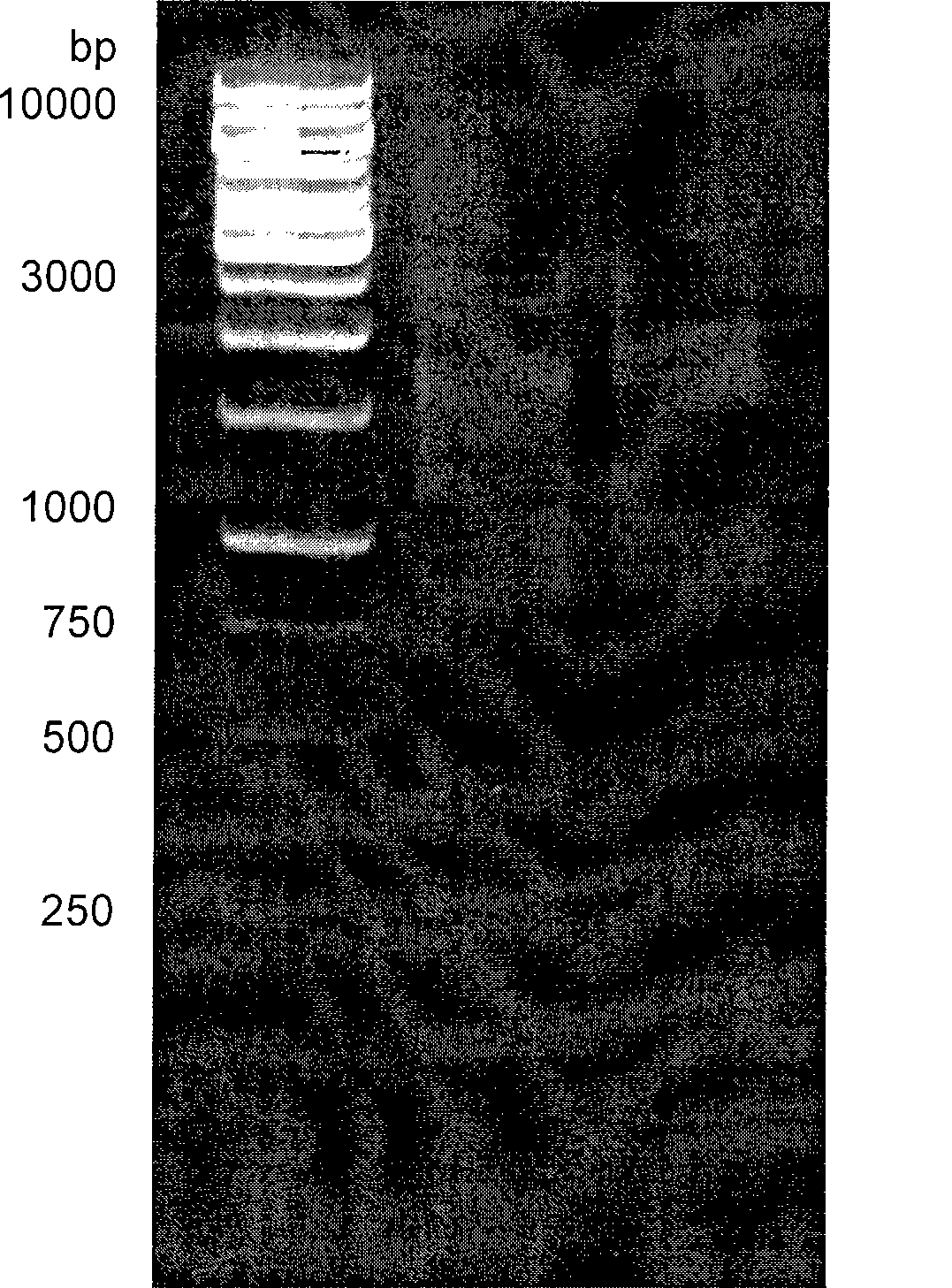
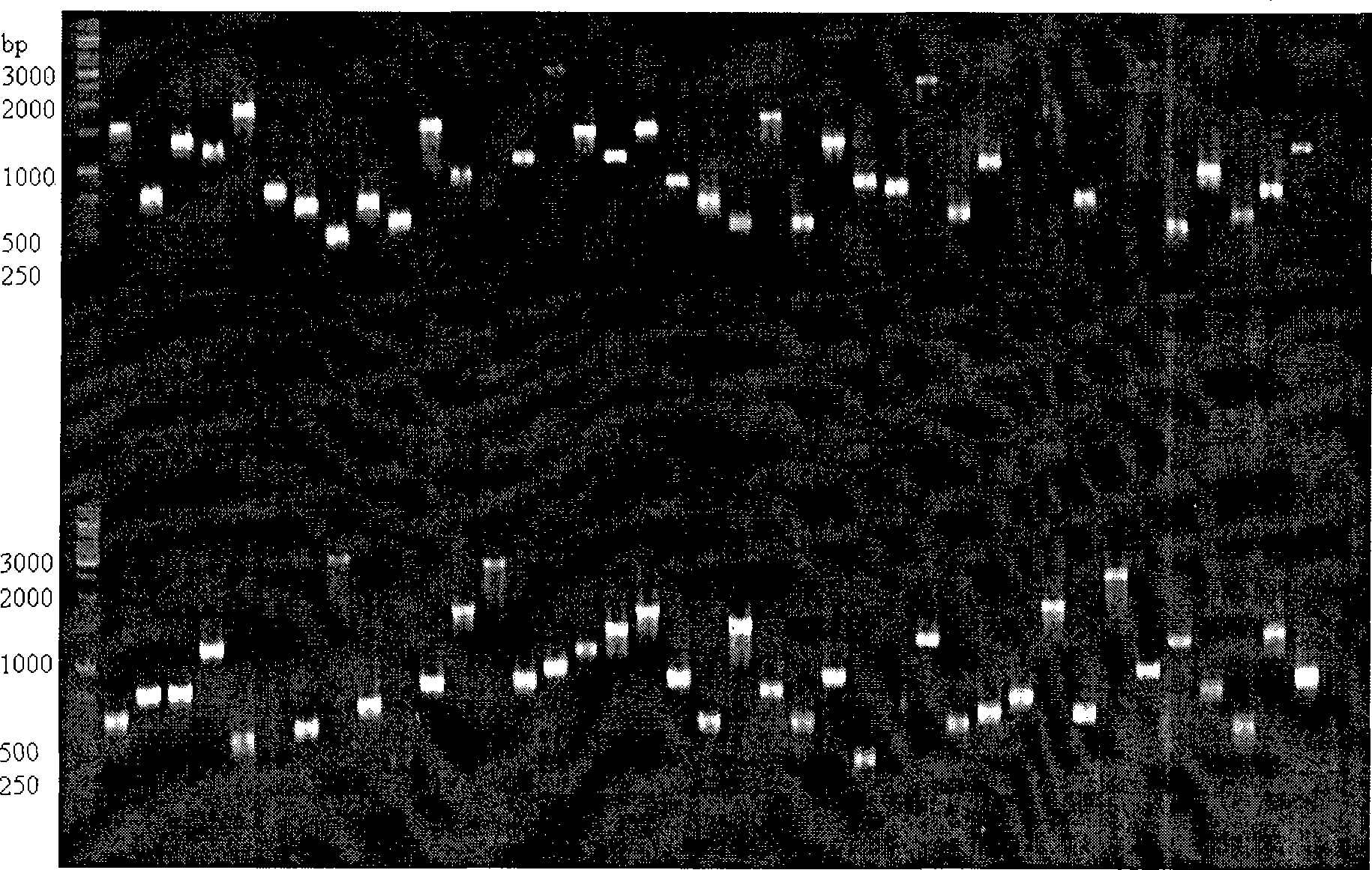
Method for constructing cDNA library
A library and sequence technology, applied in the field of cDNA library construction and molecular biology, can solve the problems of inability to easily build a library with trace precious samples, difficulty in promotion, and low success rate, and achieve a low amount of RNA and a high success rate. , the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] Construction of the cDNA library of the mangrove plant Lapis marina
[0055] Because mangrove plants grow by the sea all the year round, they have a strong ability to tolerate salt and waterlogging, and the contents in their tissues are also particularly rich, such as polysaccharides and proteins. A large number of contents will increase the difficulty and influence of extracting the RNA of mangroves. Extract the quality of RNA, so it is relatively difficult to construct a high-quality mangrove cDNA library. Using the present invention, we construct a mangrove cDNA library according to the following steps.
[0056]The requirement of the present invention for the primer of SEQ ID No.1 is: an oligodeoxynucleotide sequence, the 3' end is GGG, and the rest of the sequence is any specific linker primer sequence. The requirement of the present invention for the primer of SEQ ID No.2 is: a section of oligodeoxynucleotide sequence, 15-30 thymidine T and VN sequences at the 3' e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com