Fish transgenosis breeding method
A technology of transgenic and fish, applied to other methods of inserting foreign genetic materials, recombinant DNA technology, etc., can solve the problems of reports without in-depth research on the specific process, and achieve the effect of easy retention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
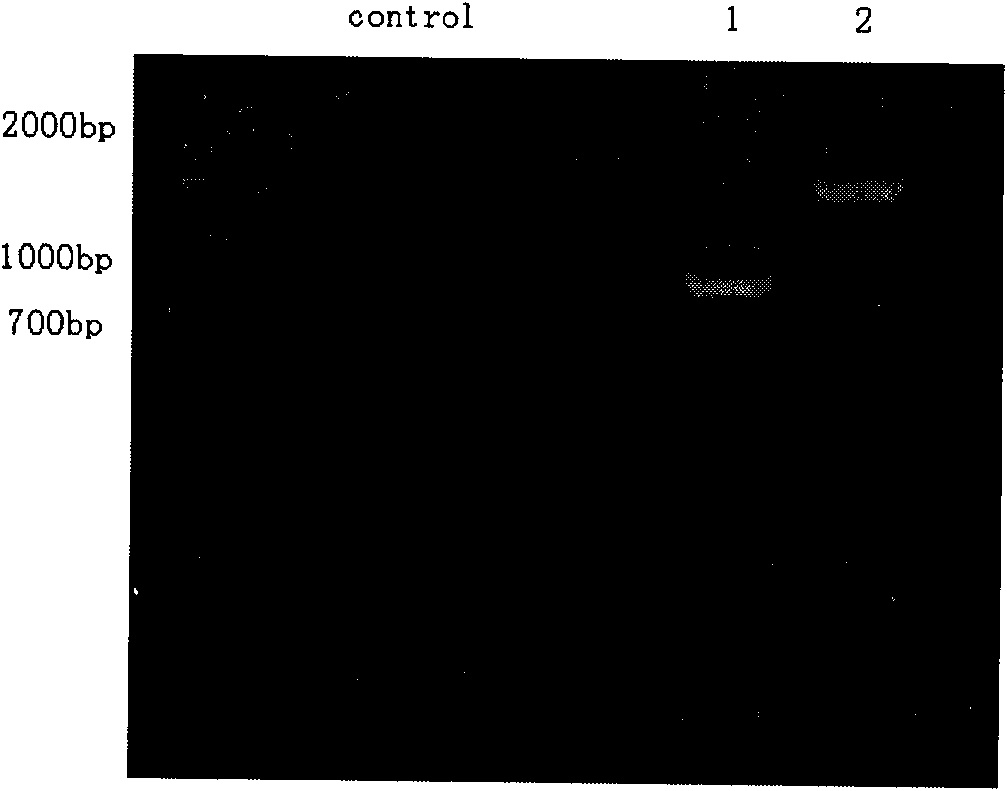
[0025] 1. Genomic DNA extraction and DNA rearrangement fragment construction:
[0026] Genomic DNA is extracted from carp (muscle or blood) (usually proteinase K method).
[0027] The extracted DNA was first digested with a restriction endonuclease Msp I (reaction system: 1-10 μg template DNA, 2U enzyme, 15 μL 10×Tango buffer in a total reaction volume of 150 μL; digestion at 37°C for 16 h), after the reaction was completed Precipitate with 2 times the volume of ethanol, centrifuge at 12,000 rpm, dry the ethanol, and reconnect with 30 μL volume (reaction system: 2.5U T4 ligase, 2 μL 10×linkagese buffer; connect overnight at 16°C), after the reaction is completed, use 2 Double the volume of ethanol precipitation, centrifuge at 12000 rpm, dry the ethanol, and then digest with the second restriction endonuclease EcoR I (reaction system: 50 μL total reaction volume containing 4U enzyme, 5 μL 10×Tango buffer; 37 Digested at ℃ for 16 h), precipitated with 2 times the volume of etha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com