Fluorescent quantitative PCR (Polymerase Chain Reaction) testing method for cccDNA (covalently closed circular deoxyribonucleic acid) of hepatitis B virus and kit thereof
A hepatitis B virus, fluorescent quantitative technology, applied in the direction of fluorescence/phosphorescence, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problem of incomplete positive strands, chimeric primers or initial probes that cannot complement it Combination and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1——The method and kit thereof for the detection of hepatitis B virus cccDNA fluorescent quantitative PCR
[0026] 1. Composition of cccDNA fluorescent quantitative PCR diagnostic kit for hepatitis B virus:
[0027] A pair of specific primers PHBV, a TaqMan fluorescent probe FPHBV, Taq enzyme, Plasmid-Safe ATP-dependent DNase enzyme, hepatitis B virus cccDNA positive control, negative control, hepatitis B virus cccDNA quantitative standard and PCR Buffer.
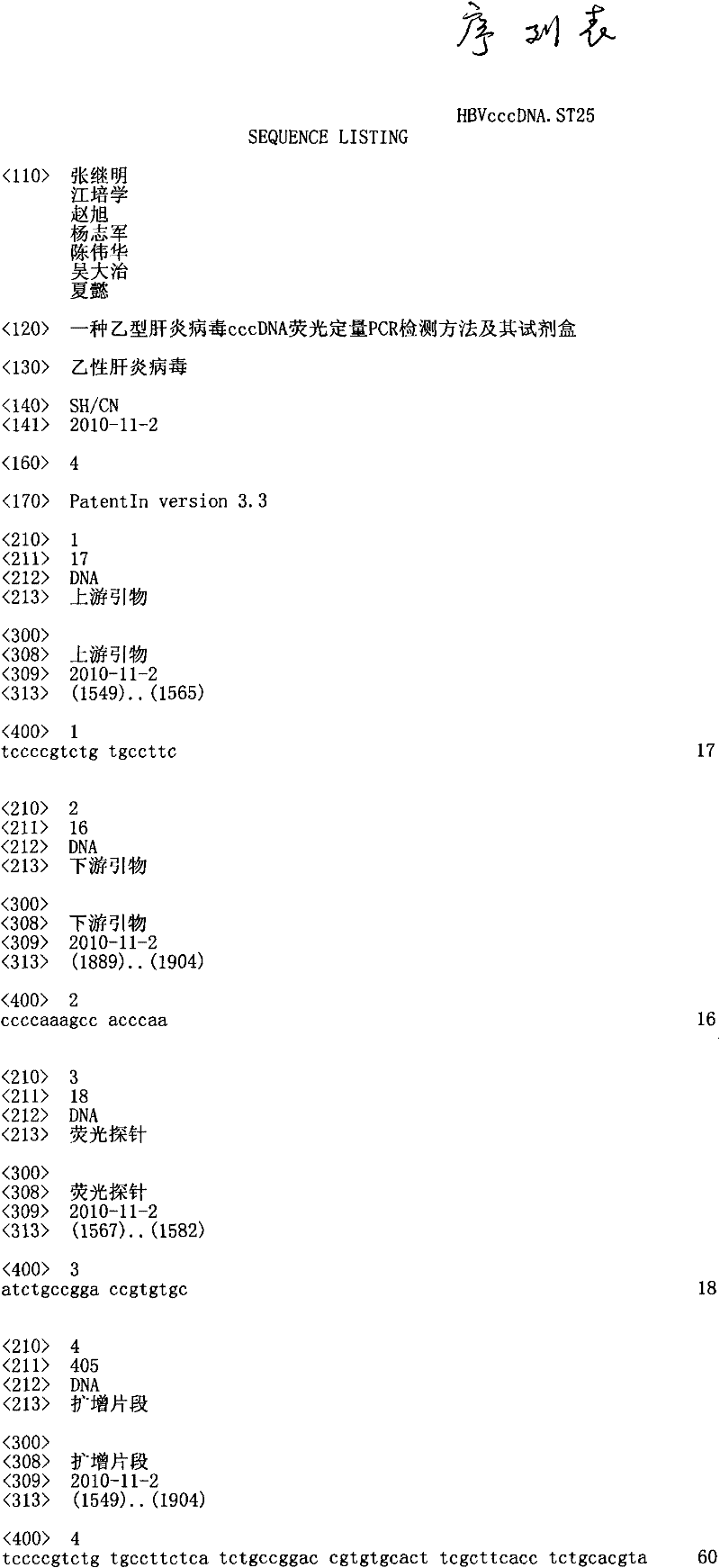
[0028] 2. According to the full sequence of HBV, in the front C coding region of the HBV genome, use Beacon Designer2.1 software to design and synthesize primers PHBV-S, PHBV-AS and probe FPHBV with the following sequences:
[0029] Primer PHBV-S:
[0030] 5'-TCCCCGTCTGTGCCTTC-3'(nt1549-nt1565)
[0031] Primer PHBV-AS:
[0032] 5'-CCCCAAAGCCACCCAA-3'(nt 1904-nt 1889)
[0033] Probe FPHBV:
[0034] 5'-FAM-ATCTGCCGGACCGTGTGC-TAMMRA-3'(nt1567-nt1582)
[0035] 3. Preparation of reaction solution: Each rea...
Embodiment 2
[0040] Embodiment 2——clinical testing
[0041] 150 cases of positive hepatitis B clinical samples were detected by the above method, among which 141 cases of HBVcccDNA were detected, accounting for 94% of the total, and the accurate quantitative analysis of HBVcccDNA was far superior to the general method. The invention has the advantages of good specificity, high sensitivity, accurate quantification, rapidity and convenience, and good repeatability. The invention designs primers spanning two gaps, so that rcDNA cannot be amplified, while cccDNA can be selectively amplified. In addition, the PSAD enzyme is used to process the extracted sample DNA, which can effectively reduce the non-cccDNA amplification and further improve the sensitivity and specificity of detection. The invention can not only be used for the detection of HBVcccDNA, but also can be used as an auxiliary diagnosis method for HBV infection in a clinical laboratory and a monitoring means for clinical treatment ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com