Method for clustering single nucleotide polymorphism (SNP) data
A clustering method and data technology, applied in the computer field, can solve problems such as inappropriate classification of data, and achieve the effects of convenient and efficient addition and deletion operations, reduction of processing efficiency, and improvement of execution efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
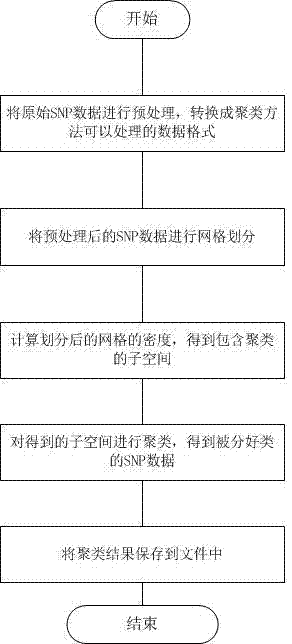
[0041] see figure 1 , this clustering method for SNP data, is characterized in that:
[0042] A. Preprocess the original SNP data and convert it into a data format that can be processed by the clustering method;
[0043] B. Mesh the preprocessed SNP data;
[0044] C. Calculate the density of the divided grid to obtain the subspace containing the clusters;
[0045] D. Cluster the subspaces obtained in step C to obtain the classified SNP data;
[0046] E. Save the clustering results to a file.
Embodiment 2
[0048] This embodiment is basically the same as Embodiment 1, and the special features are as follows:
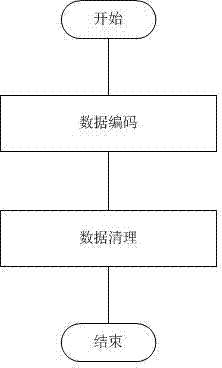
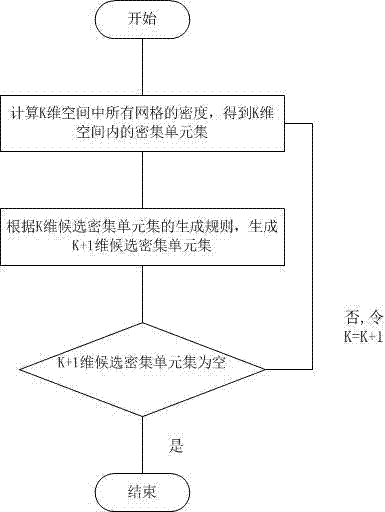
[0049] see Figure 2 ~ Figure 4 , in the step A, the original SNP data is preprocessed, and the operation steps of converting into a data format that can be processed by the clustering method are as follows:
[0050] A1) Data coding: The data format derived from SNP chip detection is as follows. Each SNP site is a typing result. There are four typing results in total, which are wild homozygous AA, mutant heterozygous AB, Mutation homozygous BB and genotyping failure mark NC; SNP data AA is coded as 0, AB is coded as 1, and BB is coded as 2;
[0051] A2) Data cleaning: if a whole row of data is NC, then the whole row of data will be deleted; if there are several NC data in a row, these NC data will be replaced with the same position of the next sample Data value; if there is more than 10% of NC data in a row, the entire row of data will be deleted.
[0052] The...
Embodiment 3
[0070] refer to Figure 1 ~ Figure 4 , a kind of clustering method for SNP data of the present invention, take the SNP data clustering of hypertensive patients as an example, its specific steps are as follows:
[0071] (1) Preprocess the original SNP data and convert it into a data format that can be processed by the clustering method, such as figure 2 As shown, the specific steps are as follows:
[0072] a) Data coding: The data format derived from SNP chip detection is as follows. Each SNP site is a typing result. There are four typing results in total, which are wild homozygous AA, mutant heterozygous AB, Mutation homozygous BB and genotyping failure mark NC; SNP data AA is coded as 0, AB is coded as 1, and BB is coded as 2;
[0073] b) Data cleaning: some whole row of data is NC, then this whole row of data will be deleted, and some have several NC data in one row, then these few NC data will be replaced with the same position of the next sample Data value; i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com