Traceablility SNP marker in pig GIGYF2 gene and detection method thereof
A detection method and gene technology, applied in the direction of recombinant DNA technology, DNA/RNA fragments, etc., can solve the problems of small differences in allele distribution frequency, lack of SNP markers, and high degree of variability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] SNP marker search
[0024] (1) Construction of DNA pool (pool)
[0025] The ear tissues of 2 Duroc pigs and 2 Shennon pigs (regardless of sex) were randomly collected respectively. After DNA was extracted, the DNA of 4 individuals was aspirated and put into the same centrifuge tube, and mixed well for use.
[0026] (2) Primer design
[0027] Using the cDNA sequence of the pig GIGYF2 gene (Genbank: NW 001885690) as a template, design primers to isolate the pig GIGYF2 gene (using the DNA of a Shannon pig as a template for PCR amplification), the primers are as follows:
[0028] Forward primer: 5'-TTTCTCCCTAAGTCGTCG-3' (see sequence SEQ ID No 2)
[0029] Reverse primer: 5'-CGTGCTGCTAAGTTTCTA-3' (see sequence SEQ ID No 3)
[0030] The total volume of the PCR reaction is 20 μl, including about 100 ng of pig GIGYF2 genomic DNA, containing 1 × buffer (Promega Company), 1.5 mmol / L MgCl2, dNTP (Shanghai Sangon Biological Company) final concentration of 150 μmol / L, and final p...
Embodiment 2
[0040] Distribution of alleles
[0041] (1) Design of test groups
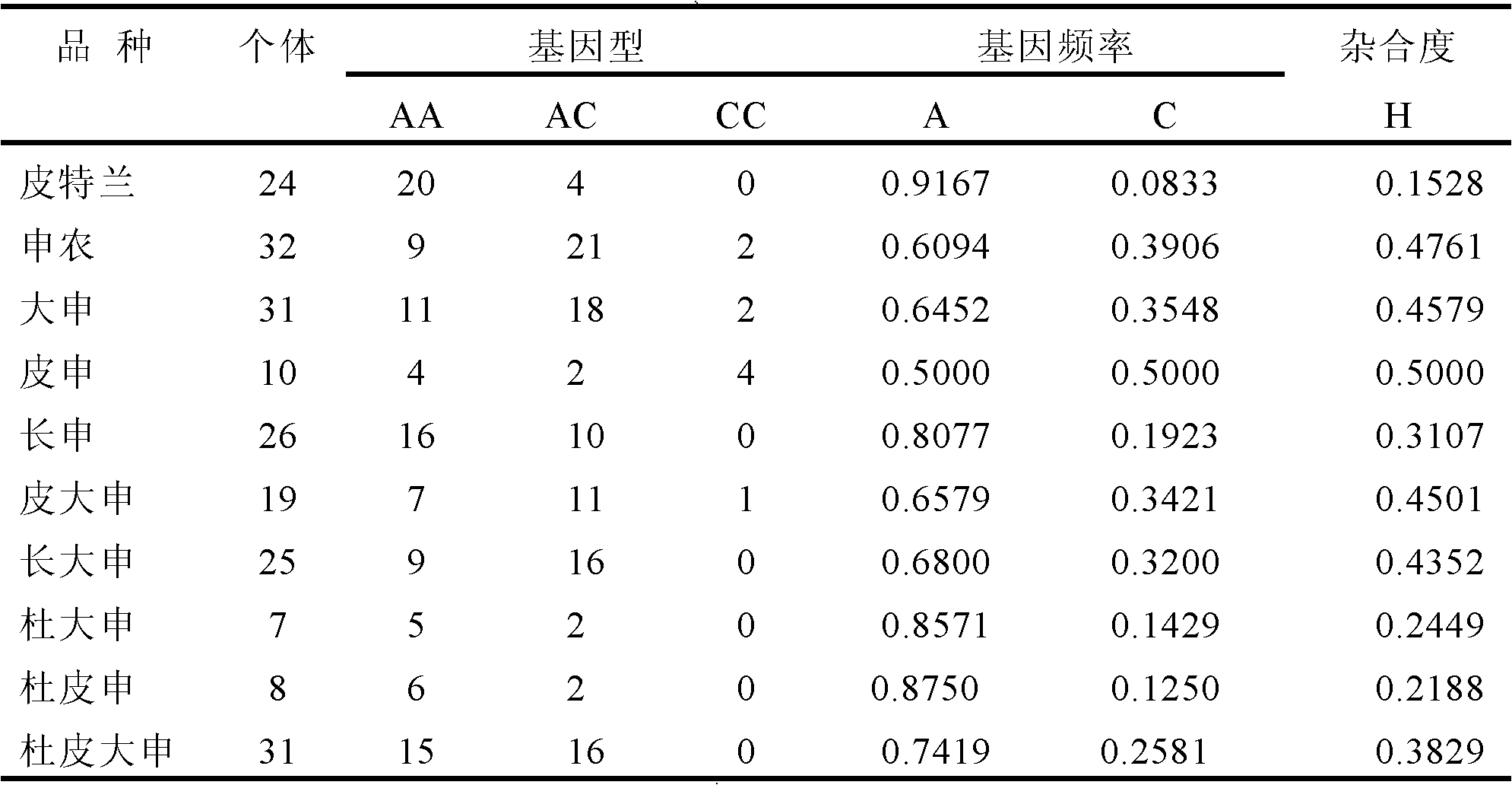
[0042] Experimental group: collected Pietrain (24 heads), Shannon (32 heads), Dashen (31 heads), Pishen (10 heads), Changshen (26 heads), Pi Dashen (19 heads), Changchang Shen (25 heads), Du Dashen (7 heads), Du Pishen (8 heads) and Du Pi Dashen (31 heads) individual ear tissues, DNA was extracted, a total of 213 DNA samples.
[0043]The purpose of the test population is to detect the distribution of SNP markers in different breeds.
[0044] (2) Genotype detection
[0045] Use forward primer: 5'-TTTCTCCCTAAGTCGTCG-3' (see sequence SEQ ID No 2)
[0046] Reverse primer: 5'-CGTGCTGCTAAGTTTCTA-3' (see sequence SEQ ID No 3)
[0047] Amplification was performed and all individual genotypes of the test population were detected by the same RFLP method.
[0048] (3) Statistical analysis
[0049] The genotypes of all individuals were recorded, and the allele frequency and heterozygosity were calculated. The result...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com