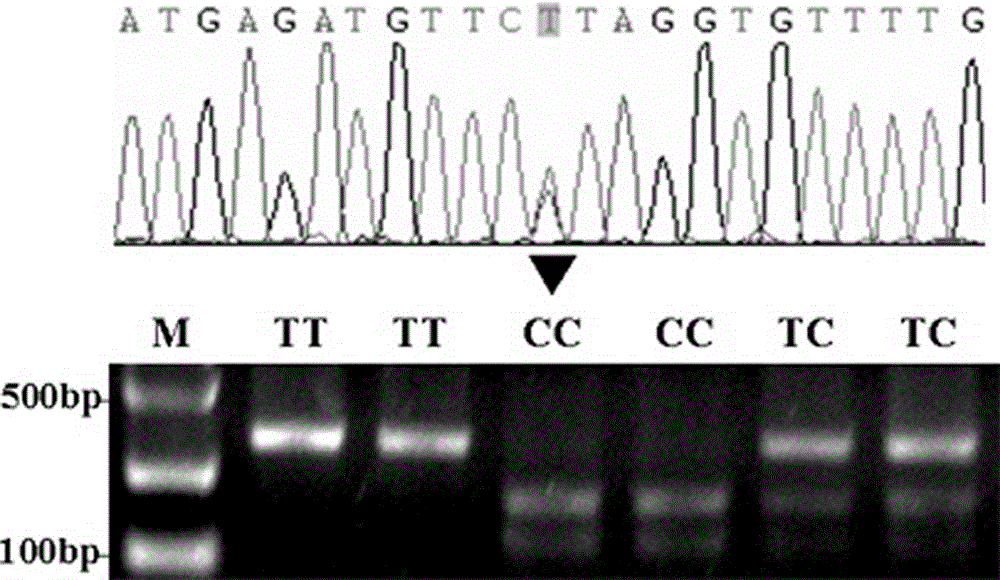
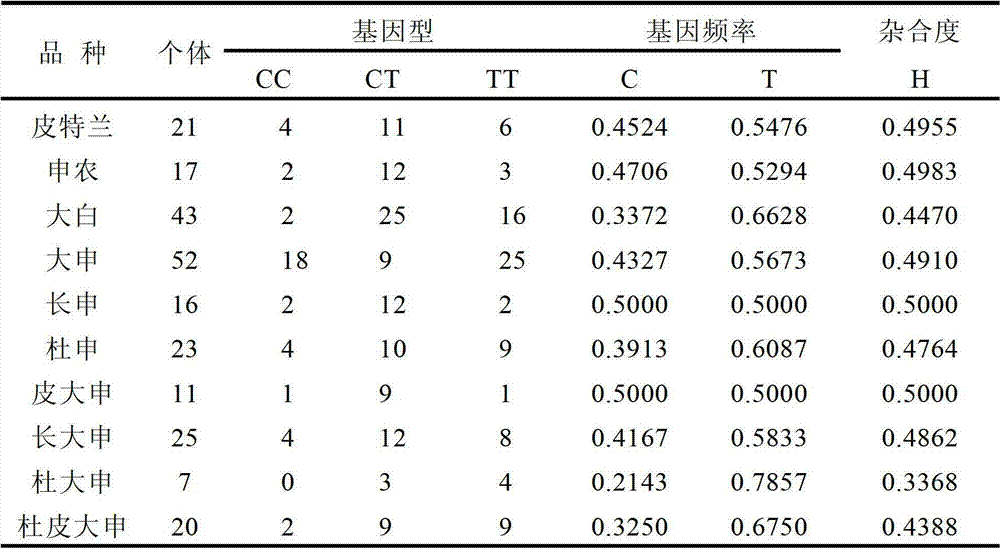
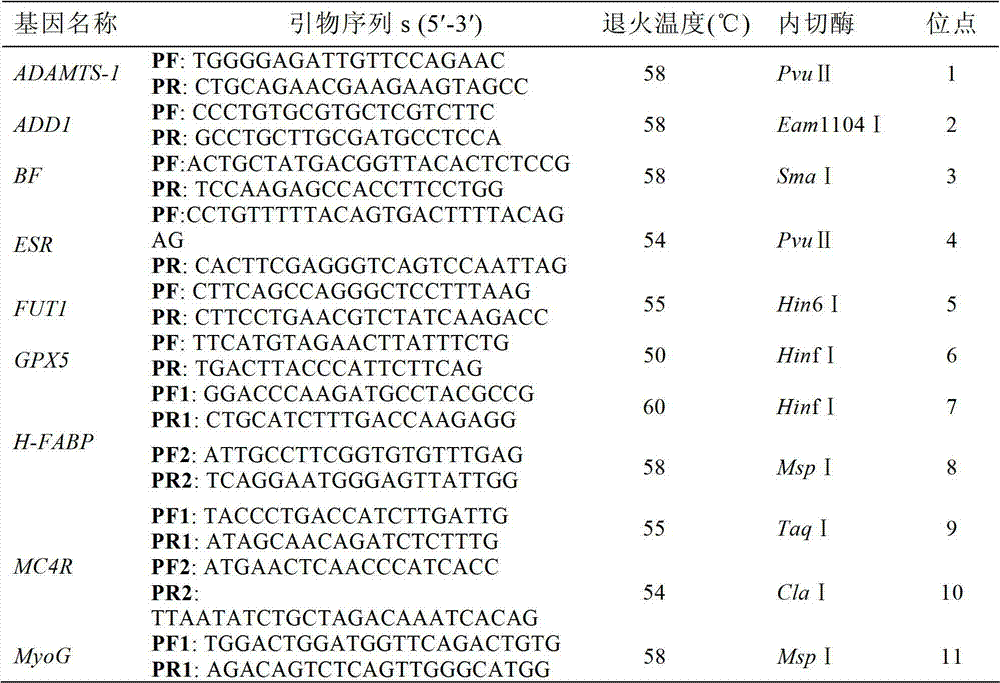
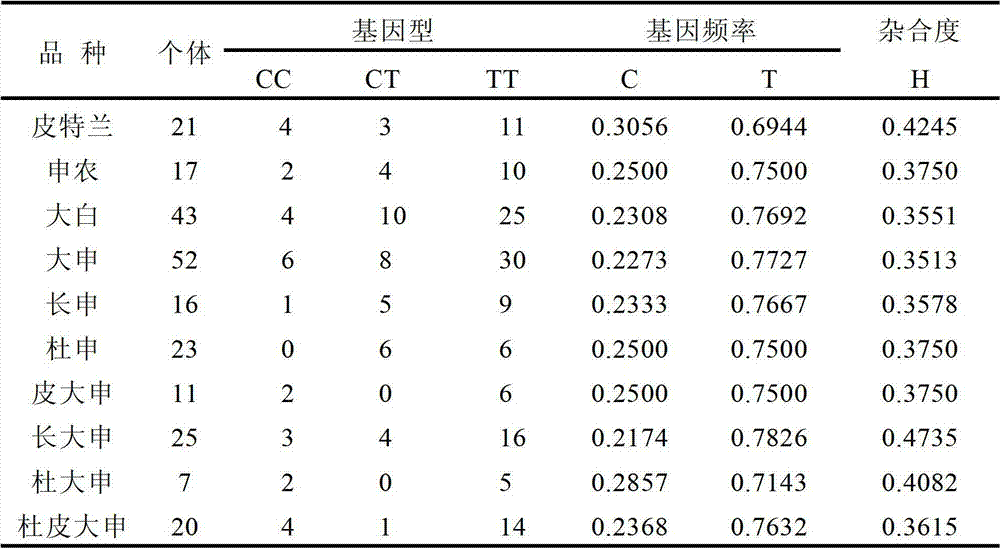
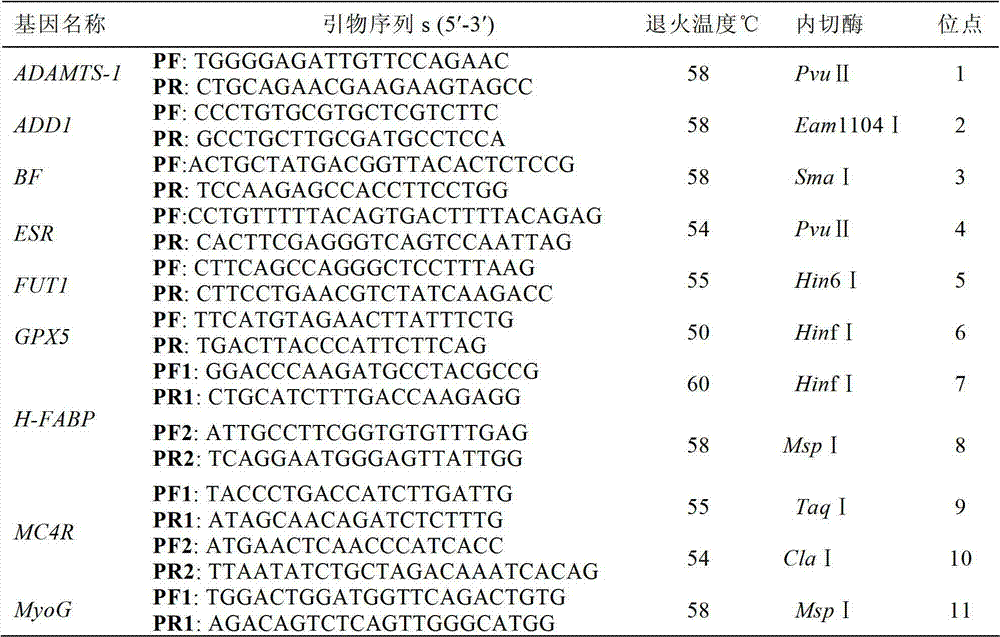
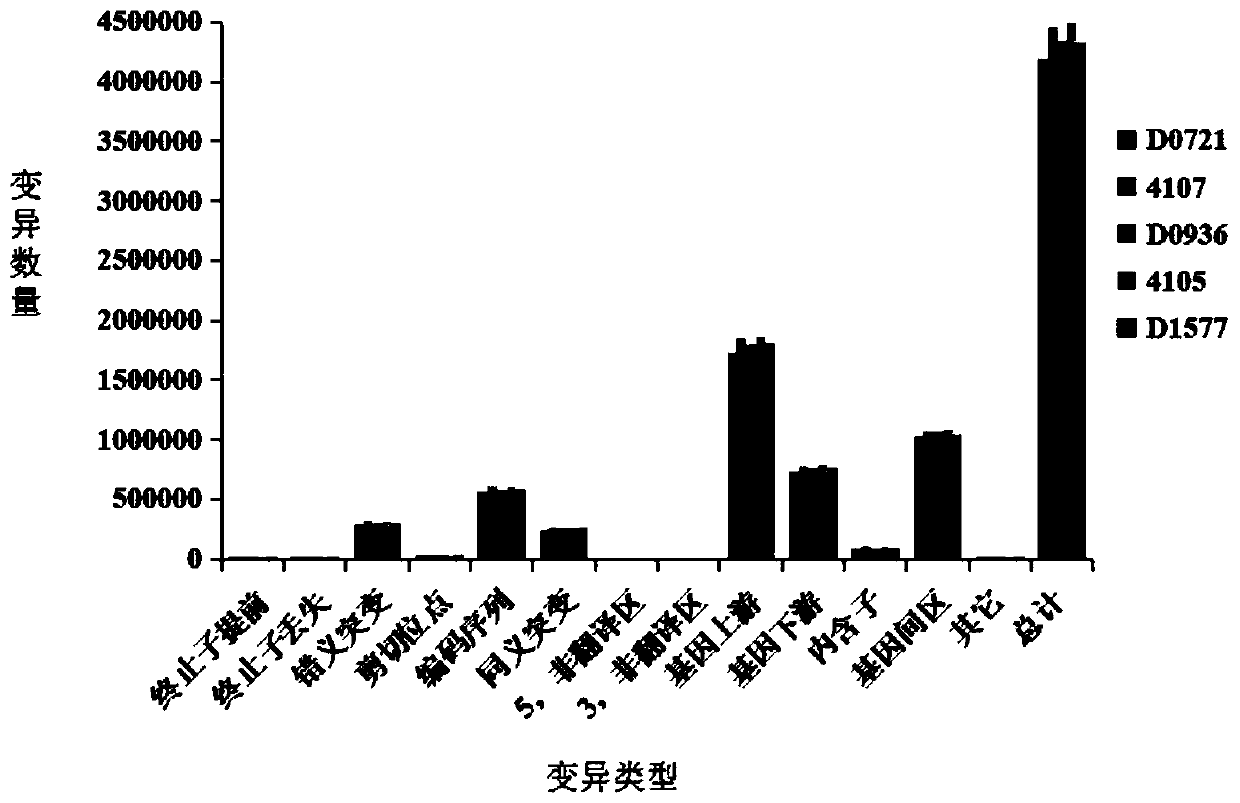
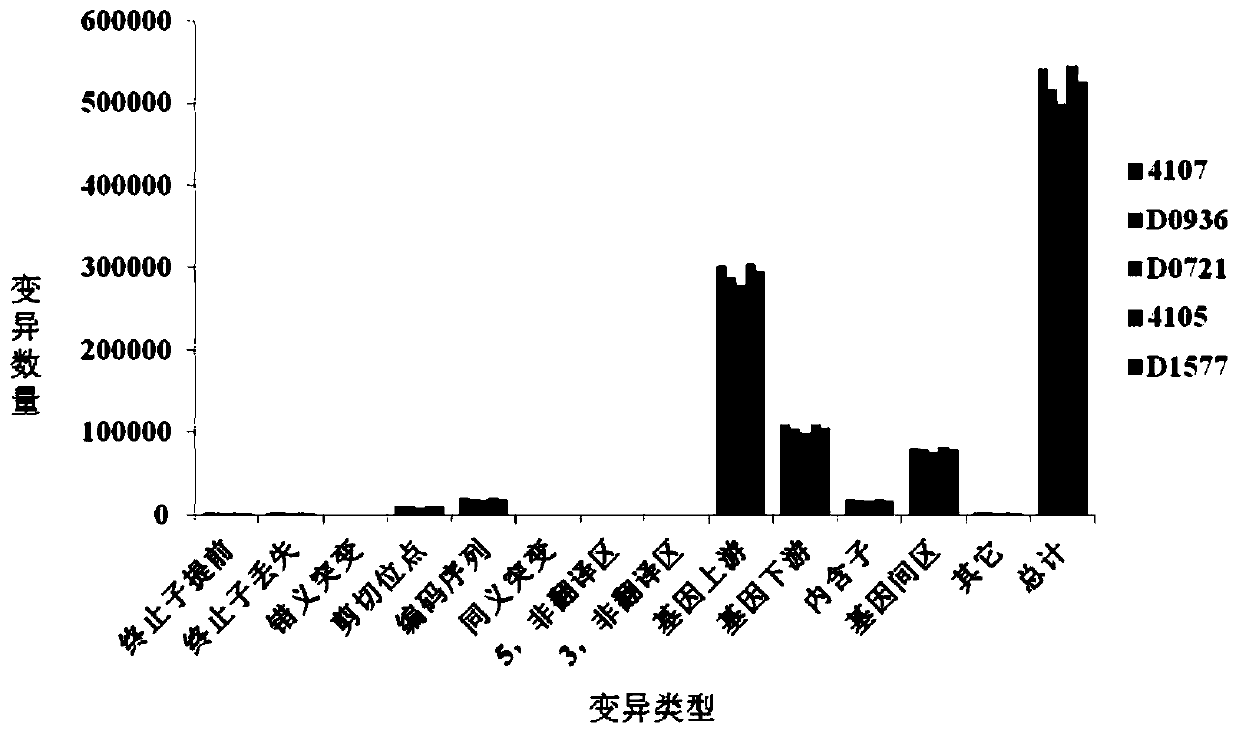
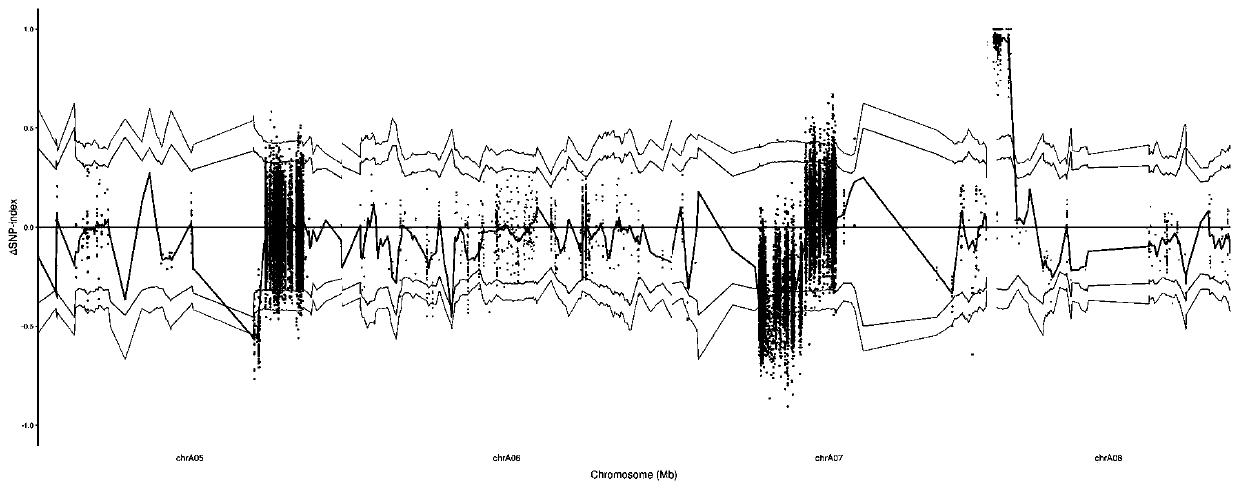
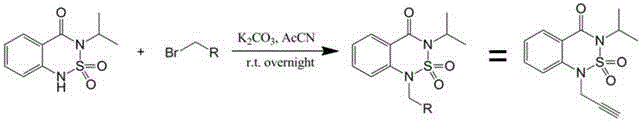
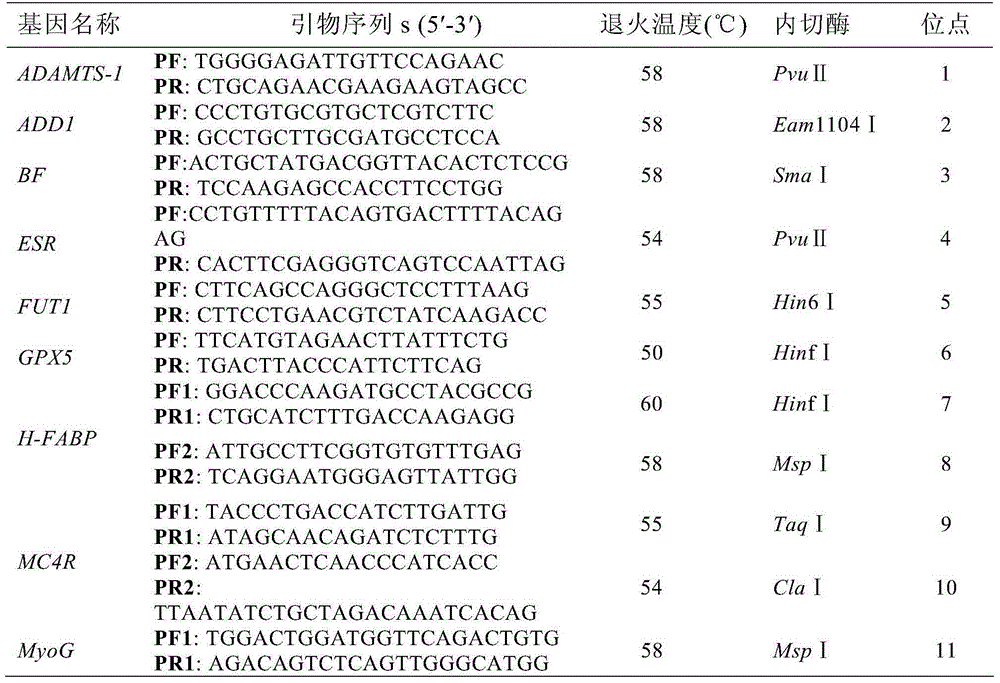
Patents
Literature
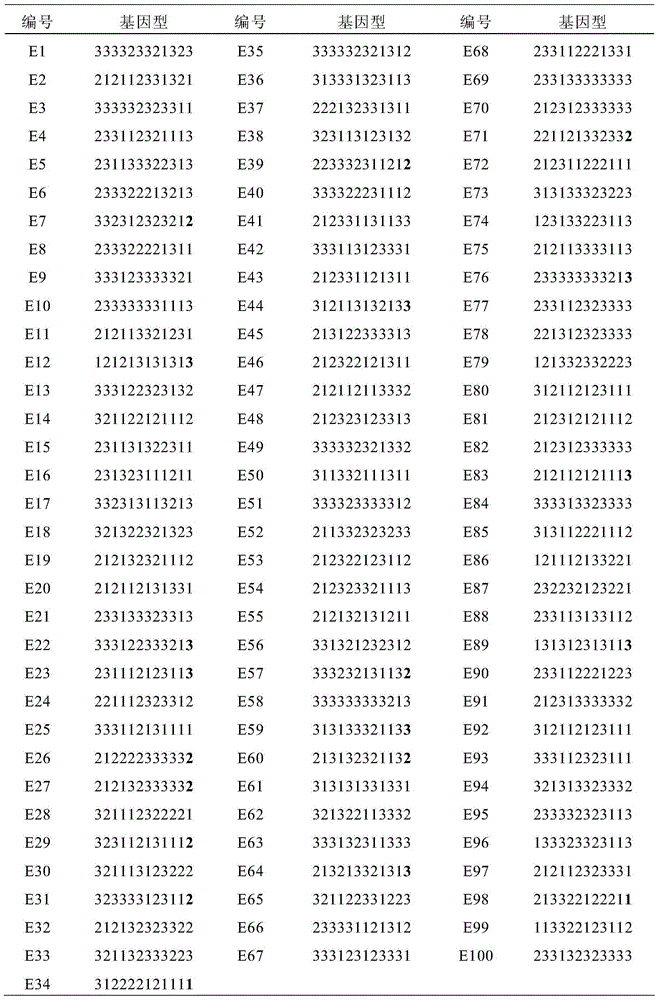
31 results about "Dna pooling" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA pooling is a practical way to reduce the cost of large-scale association studies to identify susceptibility loci for common diseases. Pooling allows allele frequencies in groups of individuals to be measured using far fewer PCR reactions and genotyping assays than are used when genotyping individuals.
In vitro selection of signaling aptamers
InactiveUS20020127581A1Bioreactor/fermenter combinationsBiological substance pretreatmentsNucleotideA-DNA
The present invention provides a method for the in vitro selection of signaling aptamers comprising the steps of synthesizing a DNA pool, the DNA having a random insert of nucleotides of a specific skewed mole ratio; amplifying the DNA pool; transcribing an RNA pool from the amplified DNA using a fluorescently labeled nucleotide; applying the fluorescently labeled RNA pool to an affinity column to remove the high-affinity fluorescent RNA molecules from the fluorescently labeled RNA pool; obtaining a cDNA pool from the high-affinity fluorescent RNA molecules; repeating the amplification and selection steps on the fluorescent RNA molecules and cloning the fluorescent RNA molecules to yield signaling aptamers. Signaling aptamers comprising DNA molecules are also selected for. Also provided is a signaling aptamer that transduces the conformational change upon binding a ligand to a change in fluorescence intensity of the signaling aptamer.
Owner:RES DEVMENT FOUND
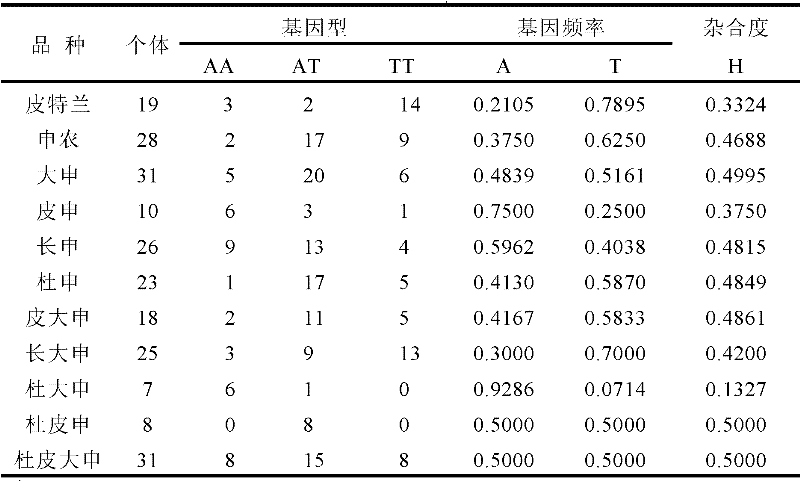
Traceablility SNP marker in pig GIGYF2 gene and detection method thereof
InactiveCN102485892AReduce distribution variancePolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNADNA fragmentation
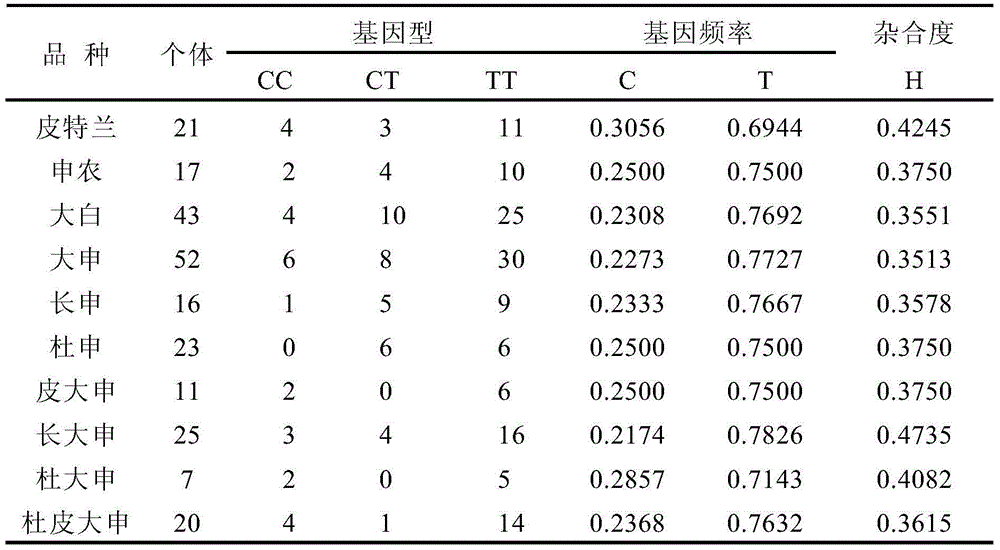
The invention relates to a traceablility SNP marker in a pig GIGYF2 gene and a detection method thereof. The SNP marker which is positioned in a genomic DNA fragment in the pig GIGYF2 gene is obtained through DNA pool sequencing. The genomic DNA fragment in the pig GIGYF2 gene, which is represented by SEQ ID NO.1 and comprises bps with the number of 452, comprises parts of twenty-three introns, and there is a base mutation from 187A to 187C at the 187th bit. The distribution case of the SNP marker of the invention in allelic genes in a test colony comprising ten pig kinds or lines is analyzed, and results show that frequencies of the marker in the allelic genes of different kinds or lines are close, so the polymorphism is abundant, there is a small difference among the frequencies of the allelic genes of different kinds or lines, and heterozygosities are greater than 0.3, so a case that the SNP marker of the invention can be used for pig product DNA traceablility is preliminarily determined.
Owner:SHANGHAI ACAD OF AGRI SCI
SRAP molecular marker closely linked with male sterility genes of tomatoes and preparation method thereof
ActiveCN104120126AGuaranteed purityImprove protectionMicrobiological testing/measurementDNA/RNA fragmentationMarker analysisGenotype
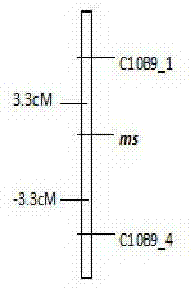
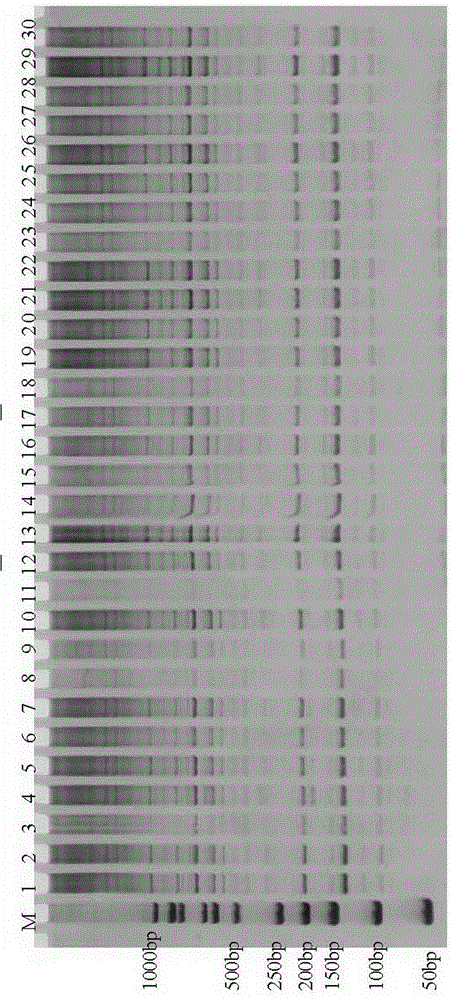
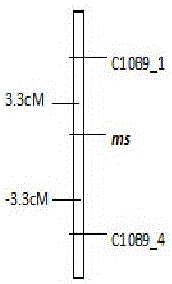
The invention discloses an SRAP molecular marker closely linked with male sterility genes of tomatoes and a preparation method thereof. The method comprises the following steps: taking purple-stem fertile tomatoes 87-5 as male parents and green-stem male sterile tomatoes at the seedling stage as female parents, hybridizing to generate F1 generation, and selfing to build an F2 segregation population; performing SRAP marker analysis on male sterility genes ms by adopting a bulked segregant analysis method, randomly combining primers before and after SRAP, selecting 544 pairs of primer combinations, and screening between male sterile and fertile pools to obtain two polymorphic markers C10B9_1 and C10B9_4 between two DNA pools; performing SRAP marker verification on the F2 segregation population by virtue of the two markers, wherein linkage analysis discovers that the linkage distance between the marker C10B9_1 and the sterility genes ms and the linkage distance between the marker C10B9_4 and the sterility genes ms are 3.3cM and -3.3cM respectively. By virtue of the molecular marker, assisted selection of male sterile tomatoes can be performed, the transfer cycle is shortened, the transfer efficiency is improved, a tedious program of identifying the sterility of each generation by selfing in a transfer process is omitted, conventional phenotype selection is transformed into genotype selection, and the accuracy and scientificity of selection are improved.
Owner:HORTICULTURE INST OF XINJIANG ACAD OF AGRI SCI
Detection method of Chinese simmental cattle carcass and meat quality trait genetic markers
ActiveCN104059963ASolve the cumbersomeFix instabilityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologySimmental cow
The invention discloses a detection method of Chinese simmental cattle carcass and meat quality trait genetic marker, and the method uses DNA pool sequencing and PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) technology for screening and detection of GPAM (glycerol-3-phosphate acyltransferase, mitochondrial) gene function mutation of Chinese simmental cattle. Through the correlation analysis, Chinese simmental cattle carcass and meat quality trait-related SNPs as a molecular genetic marker is found, and is used in the early selection and molecular pyramiding breeding of beef cattle, and breeding and improving of meat use performance of the Chinese simmental cattle can be further accelerated.
Owner:JILIN UNIV
SNP molecular marker in porcine SNCG gene for tracing and detection method thereof
ActiveCN103589716AMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceDna pools
The invention discloses a SNP molecular marker in a porcine SNCG gene for tracing and a detection method thereof. The SNP molecular marker is obtained by DNA pool sequencing and has altogether 1599 bp. The SNP molecular marker contains a part of the sequence of a third exon; a base at a position 984 mutates from 984C to 984T; the DNA sequence of the SNP molecular marker is represented by SEQ ID NO. 1. The results of analysis of the allelic distribution of the SNP molecular marker in a trial group containing 11 pig species / strains show that allelic frequencies of the SNP molecular marker in different species or strains are approximate, polymorphism is rich, allelic frequency distribution difference between species or strains is small, the degree of heterozygosis is all larger than 0.3, and thus, the SNP molecular marker disclosed herein can be used for DNA tracing for pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
SNP molecular marker in porcine AMY2 gene for tracing and detection method thereof
ActiveCN103589715AMicrobiological testing/measurementAnimals/human peptidesAgricultural scienceIntein
The invention discloses an SNP molecular marker in a porcine AMY2 gene for tracing and a detection method thereof. The SNP molecular marker is obtained through DNA pool sequencing and has altogether 1060 bp. The SNP molecular marker contains a part of the sequence of a fifth exon, a part of the sequence of a seventh exon and the complete sequences of a fifth intron, a sixth exon and a sixth intron; a base at a position 104 mutates from 104 C to 104 T; the DNA sequence of the SNP molecular marker is represented by SEQ ID NO. 1, and an amino acid sequence encoded by the SNP molecular marker is represented by SEQ ID NO. 2. The SNP molecular marker is used in cultivation of commercial pigs and is extensively applicable to DNA tracing and detection for pork products, especially to security tracing for pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
Gene fine mapping method adopting map-based cloning principle and based on plant genome sequencing and inter-subspecies hybrid segregation populations
InactiveCN108441574AReliable resultsAccurate and reliable statistical analysis resultsMicrobiological testing/measurementProteomicsWhole genome sequencingDna pools
The invention discloses a gene fine mapping method adopting the map-based cloning principle and based on plant genome sequencing and inter-subspecies hybrid segregation populations. The method comprises the following steps: (1) F2-generation segregation populations are constructed by adopting a plant I and a plant II as parents, wherein the plant I is a mutant plant with the target phenotype, andthe plant B belongs to the same species with but different subspecies from the mutant plant; (2) plants with the target phenotype are screened from the F2-generation segregation populations obtained in step (1), genomic DNA is extracted, and a specific DNA pool is obtained; whole genome sequencing is performed on the plant I, the plant II and the specific DNA pool; (3) a sequencing result obtainedin step (2) is compared with a plant referent genome; (4), positioning analysis is performed on the basis of a comparison result in step (3). The method has the advantages of accurate and reliable result, time saving, labor saving and the like and can be applied to fine mapping of plant functional genes.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
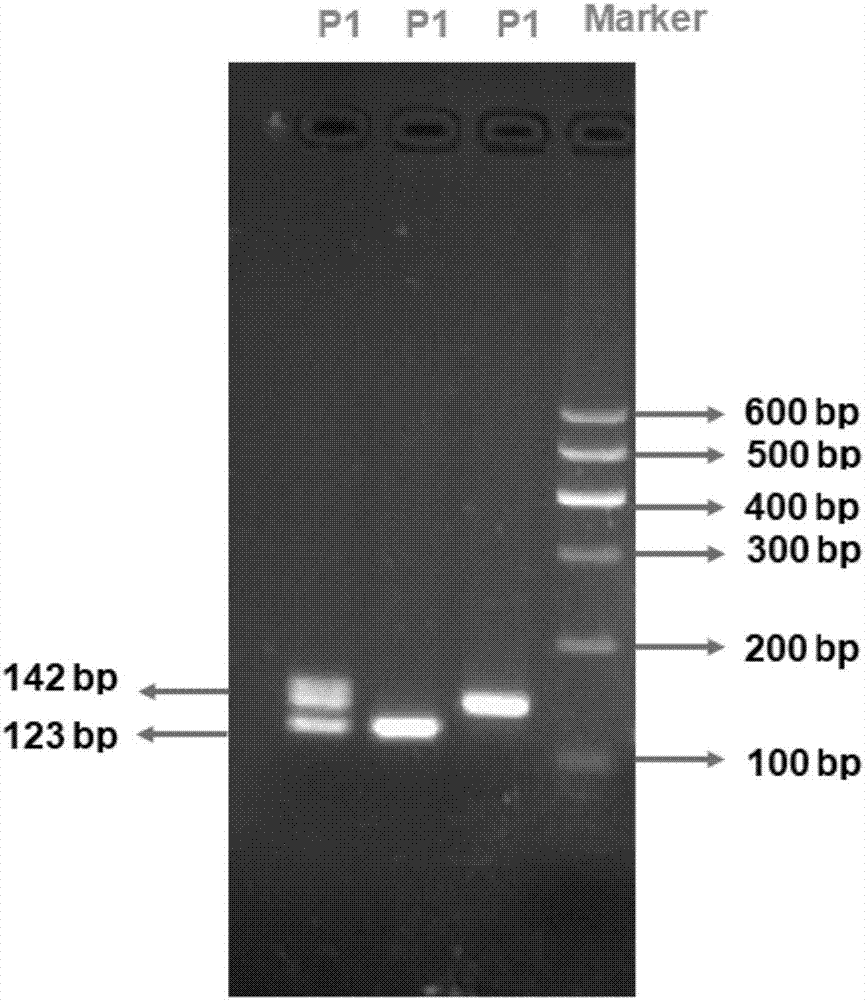
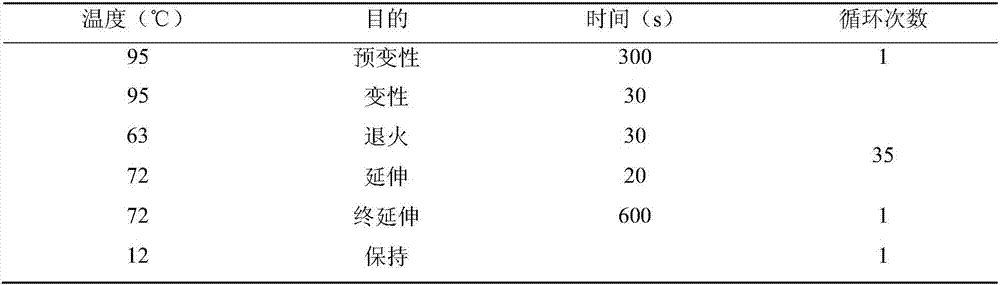
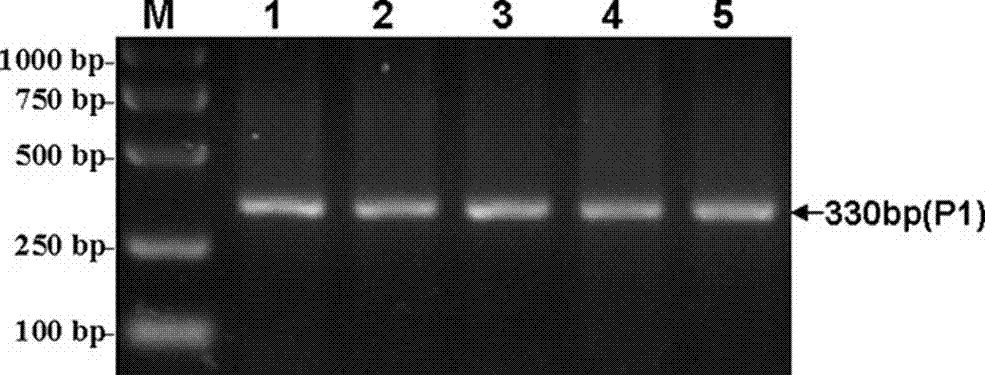
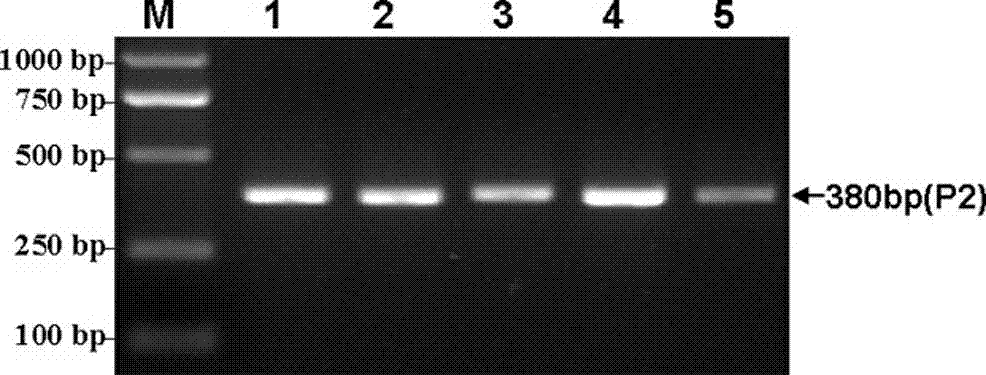
Method for detecting Indel markers of beef cattle PLAG1 genes and special kit thereof
InactiveCN107988385AImprove accuracyImprove stabilityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceDna pooling
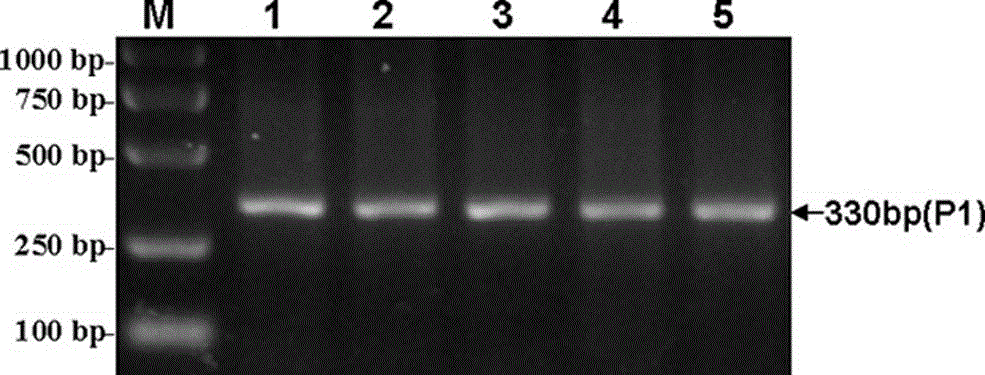
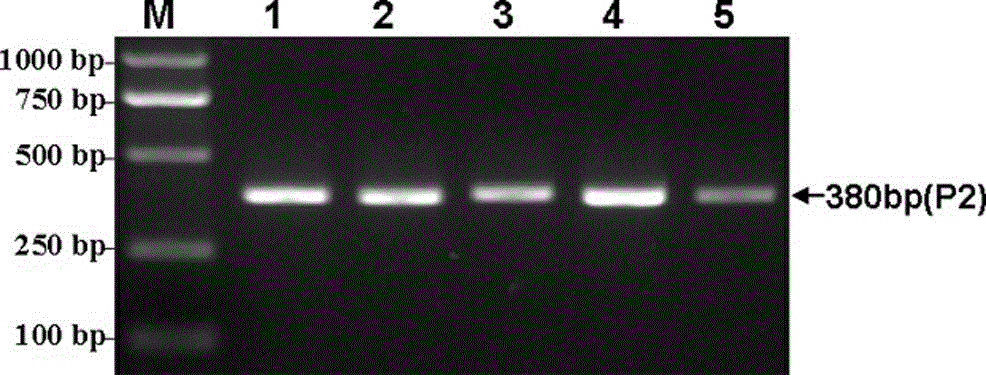
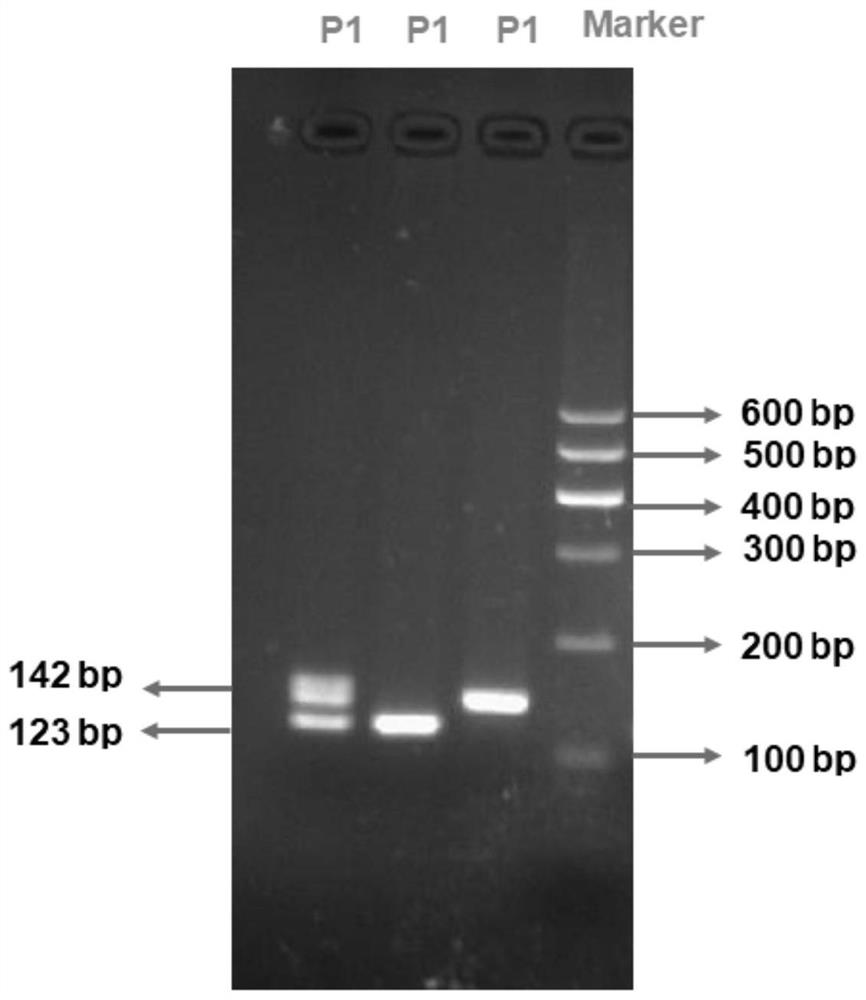
The invention discloses a method for detecting Indel markers of beef cattle PLAG1 genes and a special kit thereof. The method comprises the following steps: based on a PCR (Polymerase Chain Reaction)technology, by taking a blood genome DNA pool of beef cattle (Jiaxian red cattle) as a template, and amplifying Indel sites of cattle PLAG1 genes by utilizing a specific primer pair P1; and performingagarose gel electrophoresis, and dividing different individuals into a deletion form, a normal form and a hybrid form according to electrophoresis results. The method provided by the invention is established on a relevance basis between the Indel sites of beef cattle PLAG1 genes and growth traits, and the method is simple, rapid and convenient to popularize and use and contributes to acceleratingmolecular marker-assisted selection of the beef cattle.
Owner:NORTHWEST A & F UNIV
SNP marker used for traceability in pig SFTPC gene and detection method thereof
InactiveCN102477424APolymorphism richSmall differences in allele frequency distributionMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSFTPC gene
The invention relates to a SNP marker used for traceability in a pig SFTPC gene and a detection method thereof. The SNP molecular marker is obtained through DNA pool sequencing, and has a sequence as shown in SEQ ID NO 1; a base is mutated from 639A to 639T at the 639th base of the SNP molecular marker, and the coded amino acid sequence is shown in SEQ ID NO 2. In a test group including 11 pig varieties or strains, the allele distribution condition of the SNP molecular marker in the test group is analyzed, and the allele frequencies of the molecular marker in different varieties or strains are found to be approximate; the polymorphism is abundant; the allele frequency distribution difference between varieties or strains is fewer; the heterozygosity is more than 0.3; and the SNP marker is preliminary determined to be applicable to DNA traceablility for pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
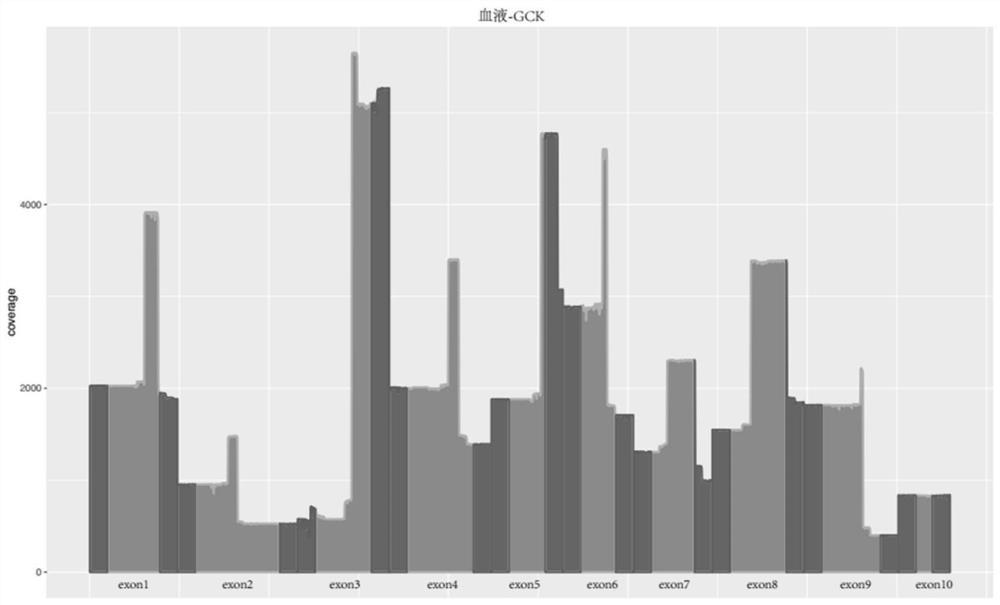
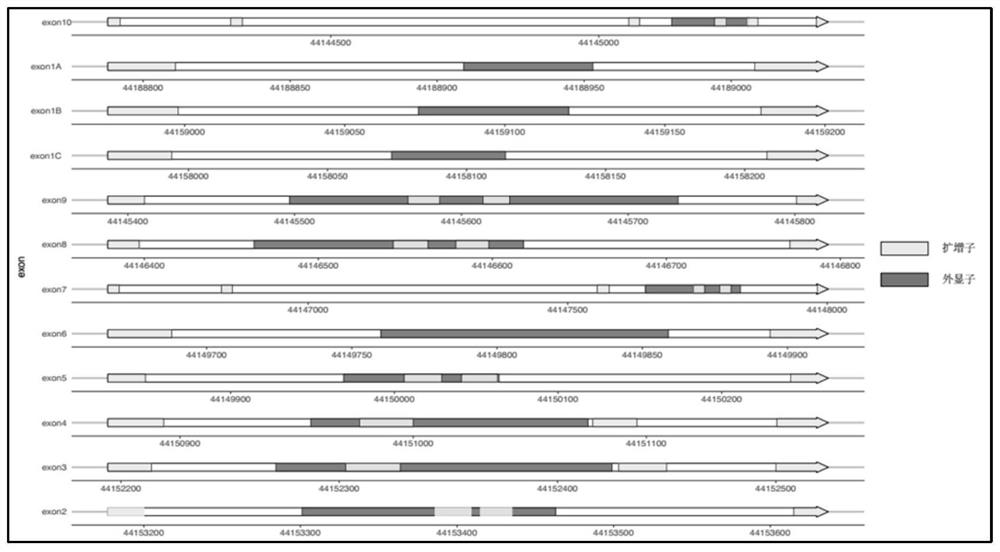
Method for detecting GCK-MODY gene mutation and kit for method
The invention provides a method for determining a quantity of DNA aggregate samples (DNA pooling) during gene detection on single-gene hereditary diseases of people. The method can be applicable to efficient analysis on a large number of samples, and the cost of gene detection on the single-gene hereditary diseases of the people can be reduced remarkably. By a primer group provided by the invention, GCK-MODY sequencing and GCK gene mutation detecting of a large number of samples can be achieved.
Owner:BIOISLAND LAB +1
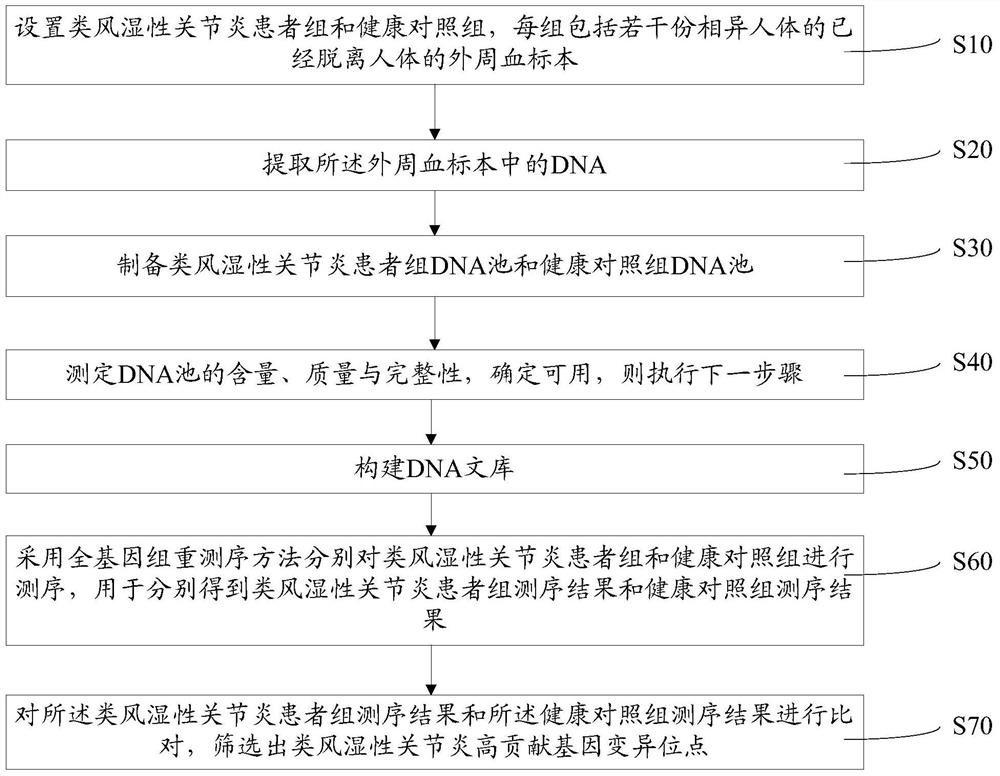
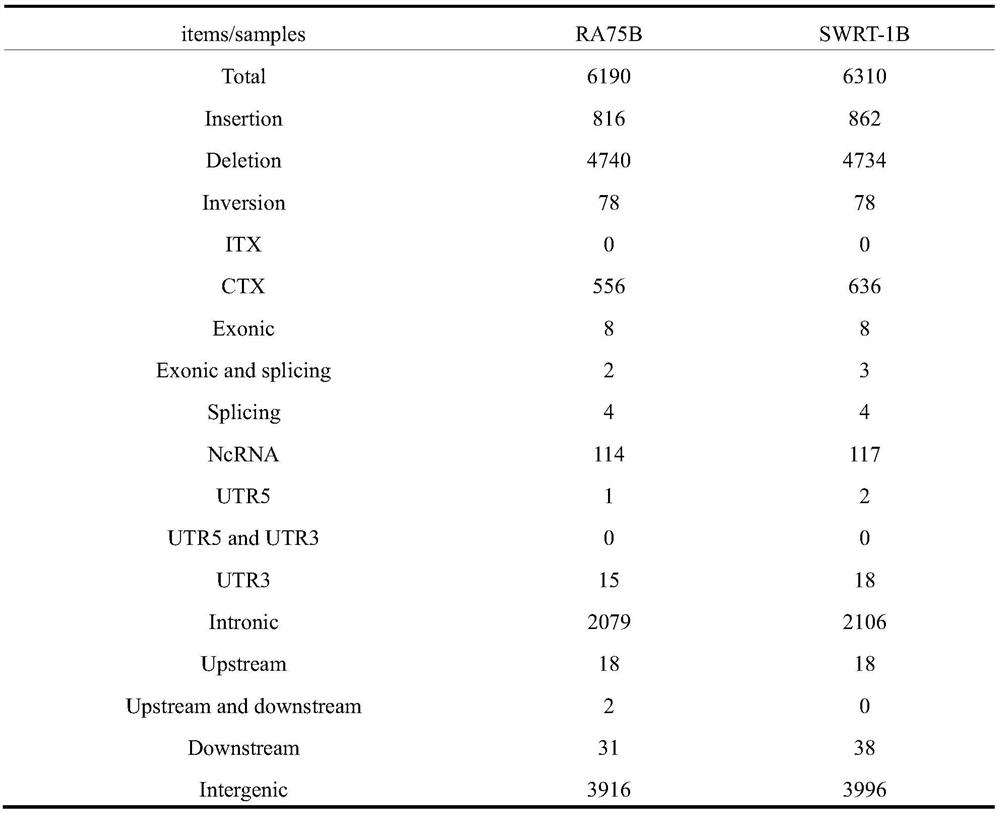
Method for screening high-contribution pathogenic gene of rheumatoid arthritis
ActiveCN107475351APlay a propelling roleMicrobiological testing/measurementHuman bodyScreening method
A method for screening a high-contribution pathogenic gene of rheumatoid arthritis comprises: setting a rheumatoid arthritis patient group and a healthy contrast group, each group comprising a plurality of peripheral-blood specimens, separated from the human body, of different human bodies; extracting DNA from the peripheral-blood specimens; preparing DNA pools of the rheumatoid arthritis patient group and the healthy contrast group; determining the content, quality and integrity of the DNA pools, and if the DNA pools are usable, carrying out the next step; building a DNA library; adopting a whole genome resequencing method to sequence the rheumatoid arthritis patient group and the healthy contrast group, and obtaining a rheumatoid arthritis patient group sequencing result and a healthy contrast group sequencing result; and comparing the rheumatoid arthritis patient group sequencing result with the healthy contrast group sequencing result, and screening high-contribution genovariation sites out. According to the method, high-contribution pathogenic genovariation sites are screened, and the method is beneficial for early stage treatment and prevention of rheumatoid arthritis.
Owner:戴勇 +1
Brassica napus wax powder gene positioning method based on whole genome resequencing
ActiveCN111575399ASequencing data is sufficientQuality standardsMicrobiological testing/measurementSequence analysisBiotechnologyGenetic correlation analysis
The invention provides a brassica napus wax powder gene positioning method based on whole genome resequencing, and belongs to the technical field of biological information. A near-isogenic line is established on the basis of a segregation population. Mixed grouping analysis is adopted, wax powder-free plants in a 3: 1 segregation population, all wax powder phenotypic insegregation populations andthree parents in a near-isogenic line are selected, and five DNA pools are built in total for genome resequencing. And genetic correlation analysis is performed by utilizing resequencing data, and a locus for controlling the character of the wax powder is positioned in a 590663-1657546bp region of an A08 chromosome. The method provided by the invention has the biggest characteristic that all offspring individuals do not need to be detected, but offsprings with two extreme characters are subjected to mixed pool analysis. Moreover, the method provided by the invention has sufficient sequencing data and qualified quality, and can be used for next analysis.
Owner:SHANGHAI ACAD OF AGRI SCI
DNA aptamer sequences for recognizing bentazone and derivatives thereof
ActiveCN106047872AWide range of environmental detection application prospectsRapid detection expansionBiological testingDNA/RNA fragmentationDNA AptamersDna pools
The invention provides DNA aptamer sequences for recognizing bentazone and derivatives thereof. DNA aptamer as referred in the invention is single-stranded oligodeoxynucleotide with different oligodeoxynucleotide sequences; and the sequences are capable of recognizing the herbicide bentazone in a solution and applicable as biological recognition materials for detection of the content of bentazone in samples. The DNA aptamer sequences are obtained through in-vitro seletion or SELEX from a random DNA pool. In an aqueous solution, the bonding strength of the DNA aptamer sequences and bentazone molecules dissociation constant enables a dissociation constant (Kd) to reach the magnitude of [mu]M.
Owner:TSINGHUA UNIV
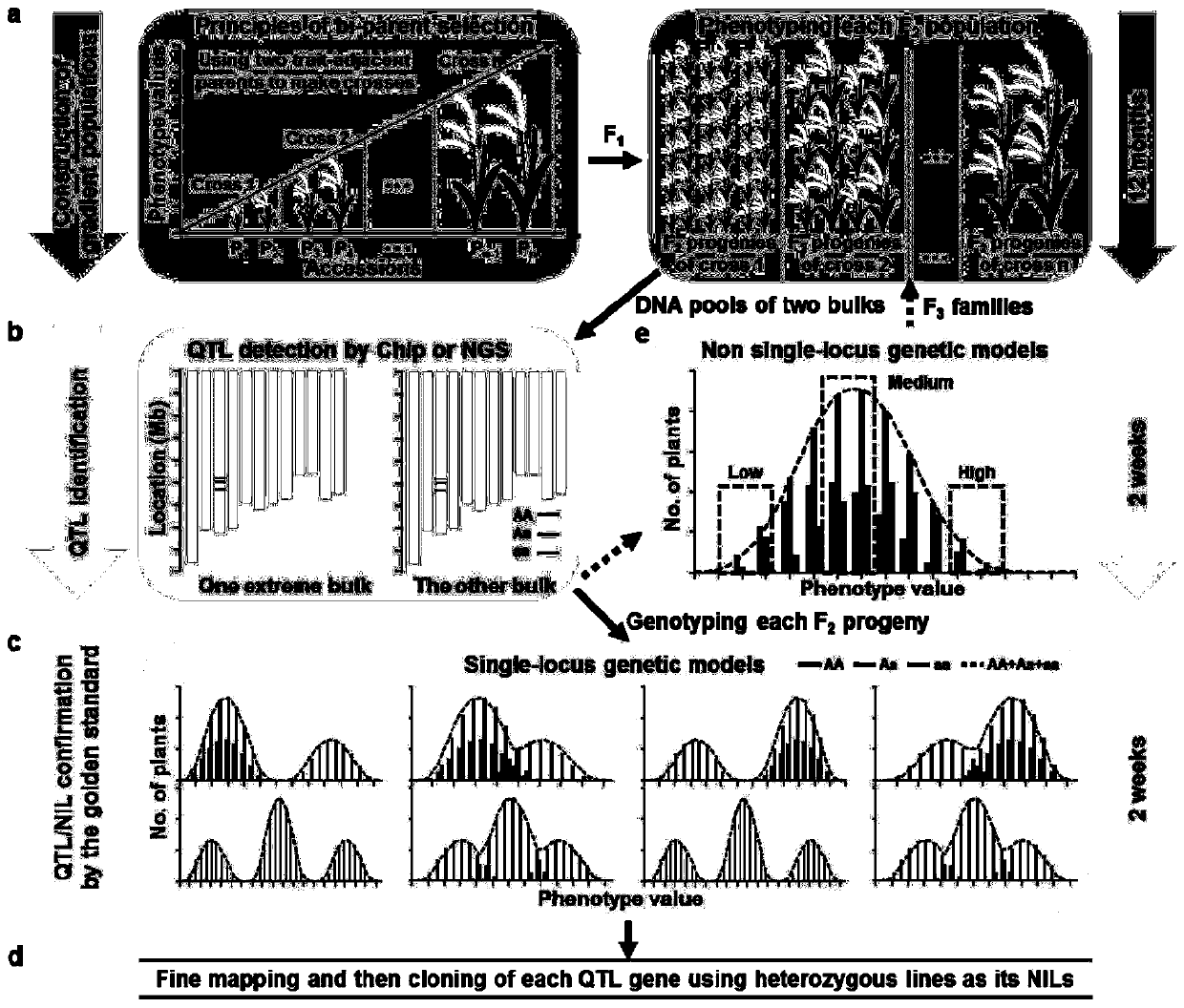
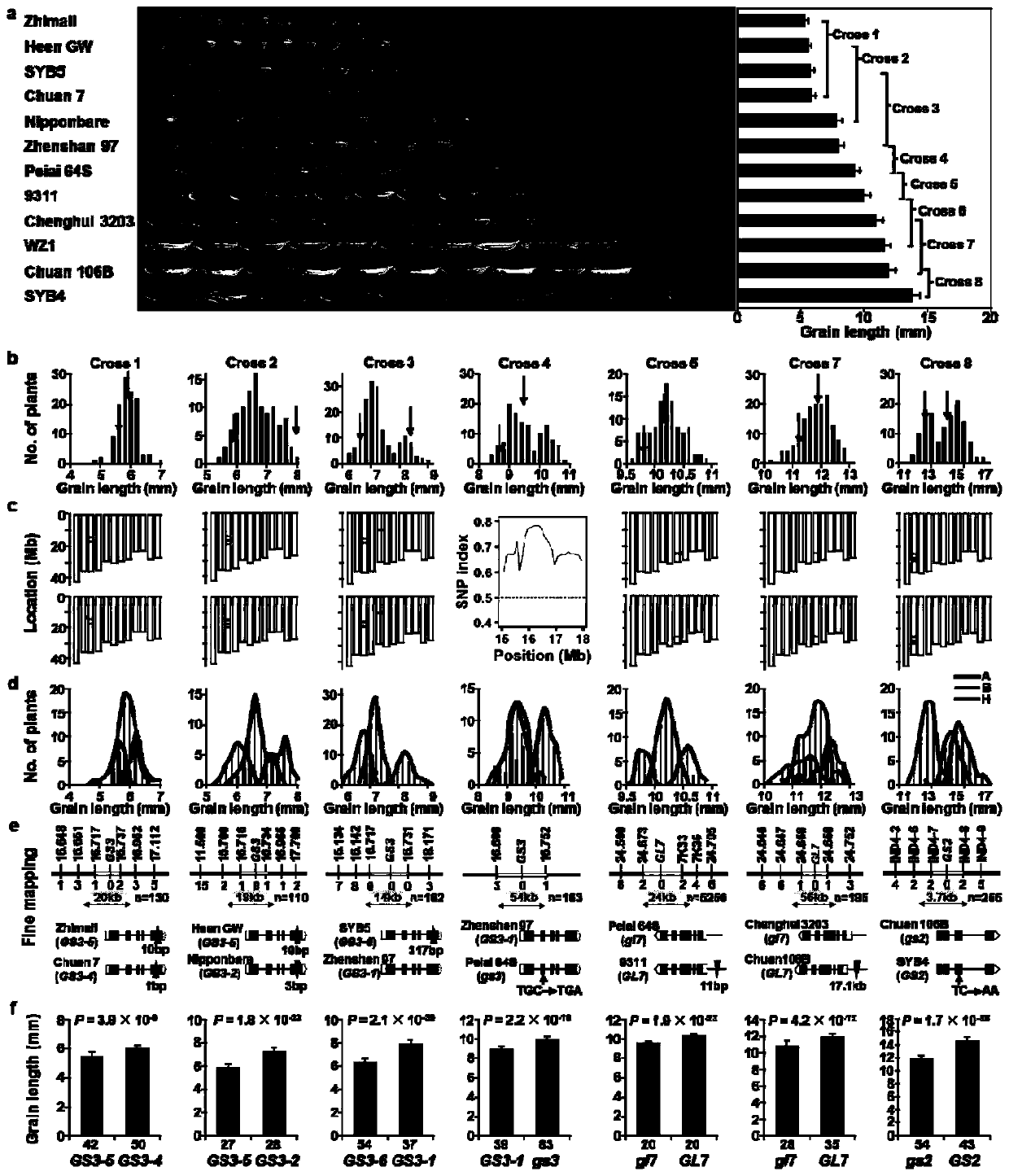
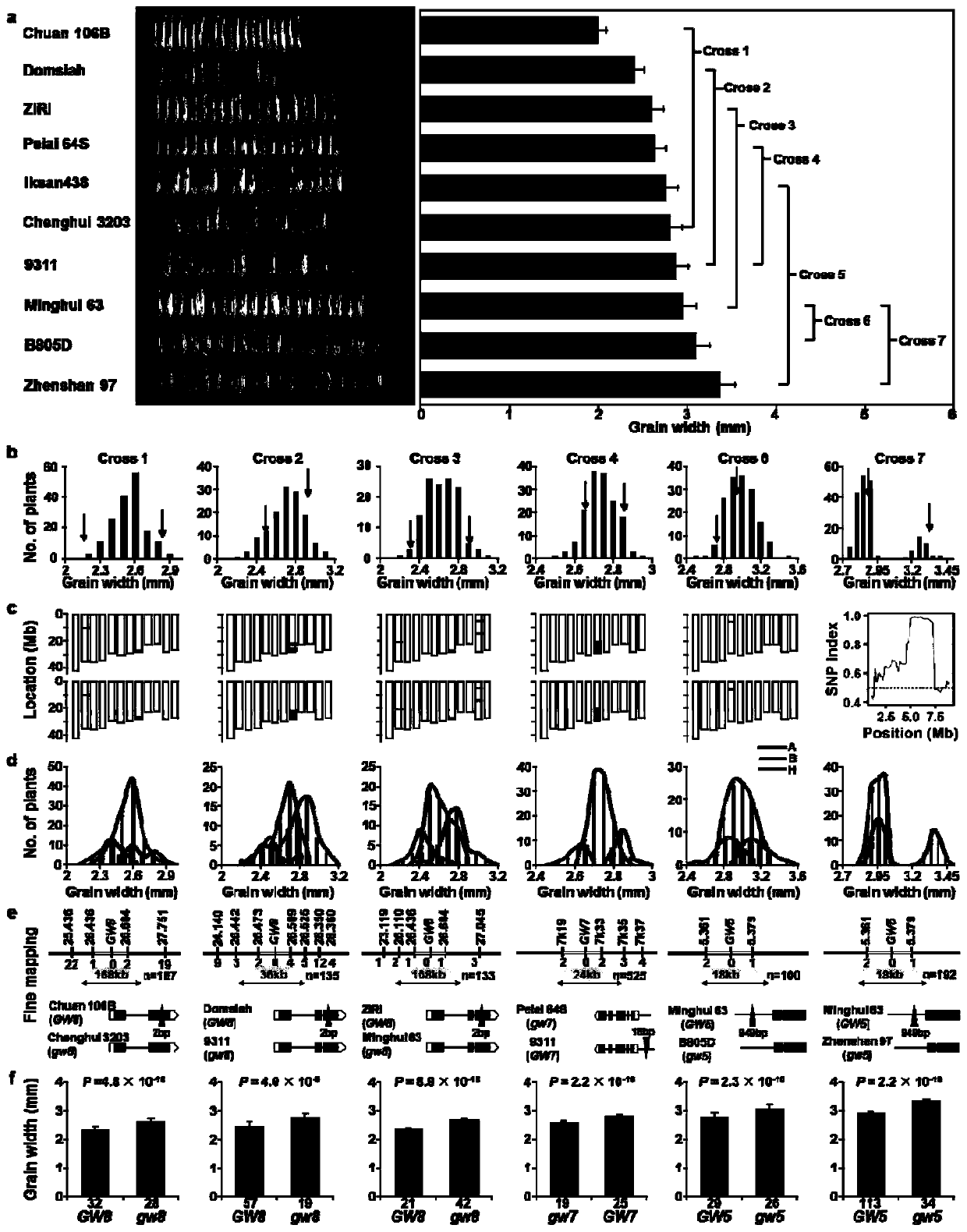
RapMap method for rapid and high-throughput positioning and cloning of plant QTL gene
The invention discloses a RapMap method for rapid and high-throughput positioning and cloning of a plant QTL gene. The method comprises the steps of: selecting strains with relatively small characterdifferences from the core germplasm as gradient parents; constructing as many F2 gradient genetic populations as possible; preliminarily positioning QTL by performing chip detection or next-generationsequencing on two separated extreme phenotypic DNA pools of each group, verifying the QTL at the single plant level of each gradient group according to the co-separation criteria provided by the invention, and cloning a target gene by using a QTL heterozygous family as a near-isogenic line for fine positioning. The concept of gradient genetic populations and the idea of co-segregation criteria are the core of the invention. Six rice grain length and grain width genes are successfully cloned by applying the RapMap method, and the RapMap is indicated to be a rapid and high-throughput gene cloning RapMap method integrating QTL positioning, verification and near-isogenic line screening into a whole.
Owner:HUAZHONG AGRI UNIV
Simplified QTL mapping approach for screening and mapping novel markers associated with beef marbling
InactiveUS20070224623A1Simplified and inexpensiveSmall sample sizeMicrobiological testing/measurementBiological testingDna poolingSelective genotyping
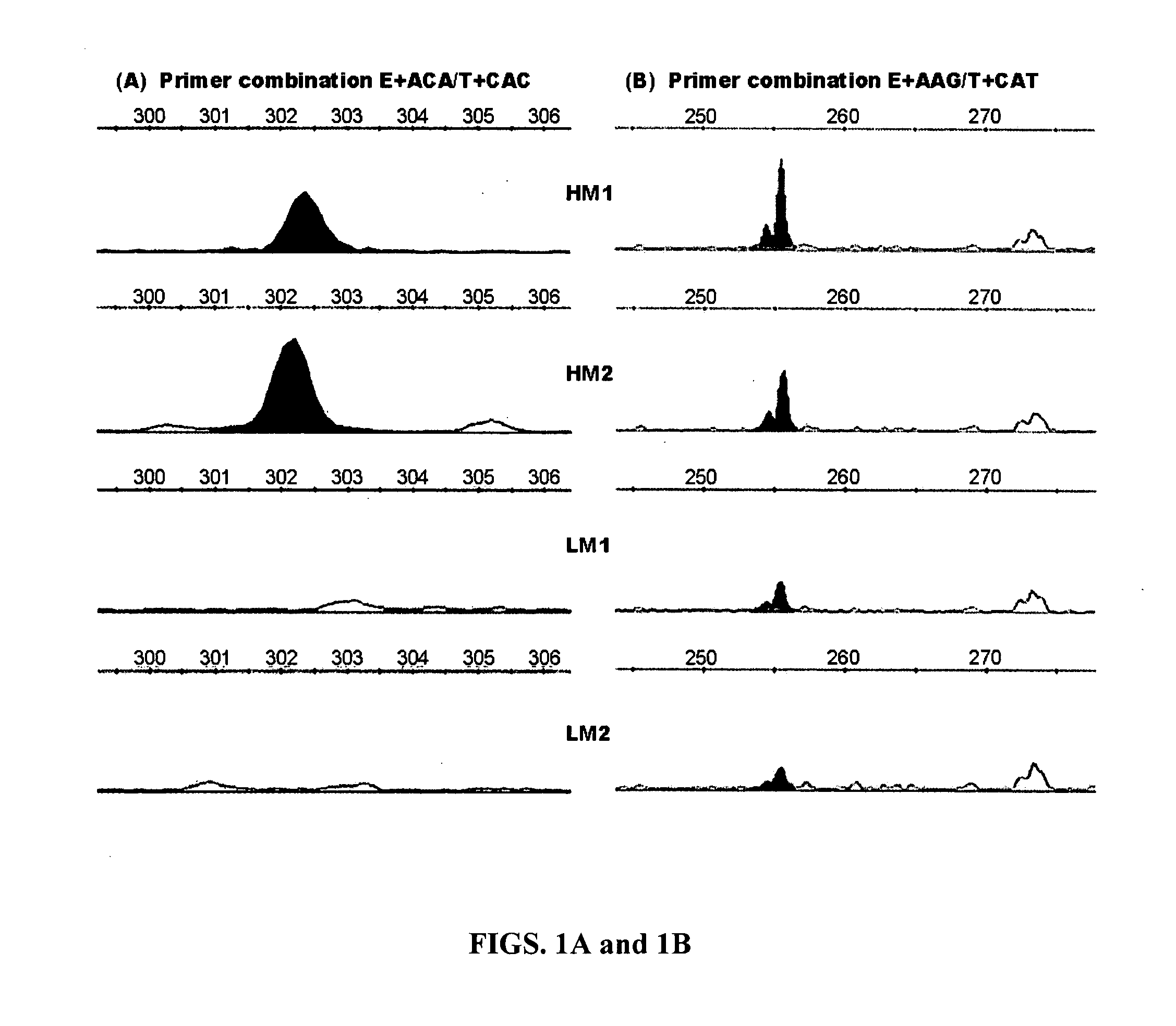
Particular aspects provide an inexpensive QTL mapping approach for genome-wide scans of QTL linked markers and for narrowed locations of QTL regions, which may comprise integration of the amplified fragment length polymorphism (AFLP) with DNA pooling and selective genotyping and comparative bioinformatics tools. AFLP simultaneously screens high numbers of loci for polymorphisms and detects many more polymorphic DNA markers than any other PCR based detection systems, and provides for identification of “responsible mutations” for a QTL. Aspects of the present invention also provide novel compositions and methods based on novel DOPEY2 and / or KIAA1462 single nucleotide polymorphisms such as AAFC03071397.1:g.12881G>C, g.12925G>A, g.12951T>C, g.13013A>G, g.13125G>A and g.13173C>T in the DOPEY2 gene and AAFC02113318.1:g.1367G>A and g.1372G>A in the KIAA1462 gene, which may provide novel markers for beef marbling and subcutaneous fat. Additional aspects provide for novel methods which may comprise marker-assisted selection to improve beef marbling and subcutaneous fat in cattle.
Owner:WASHINGTON STATE UNIV RES FOUND INC
Accurate and massively parallel quantification of nucleic acid
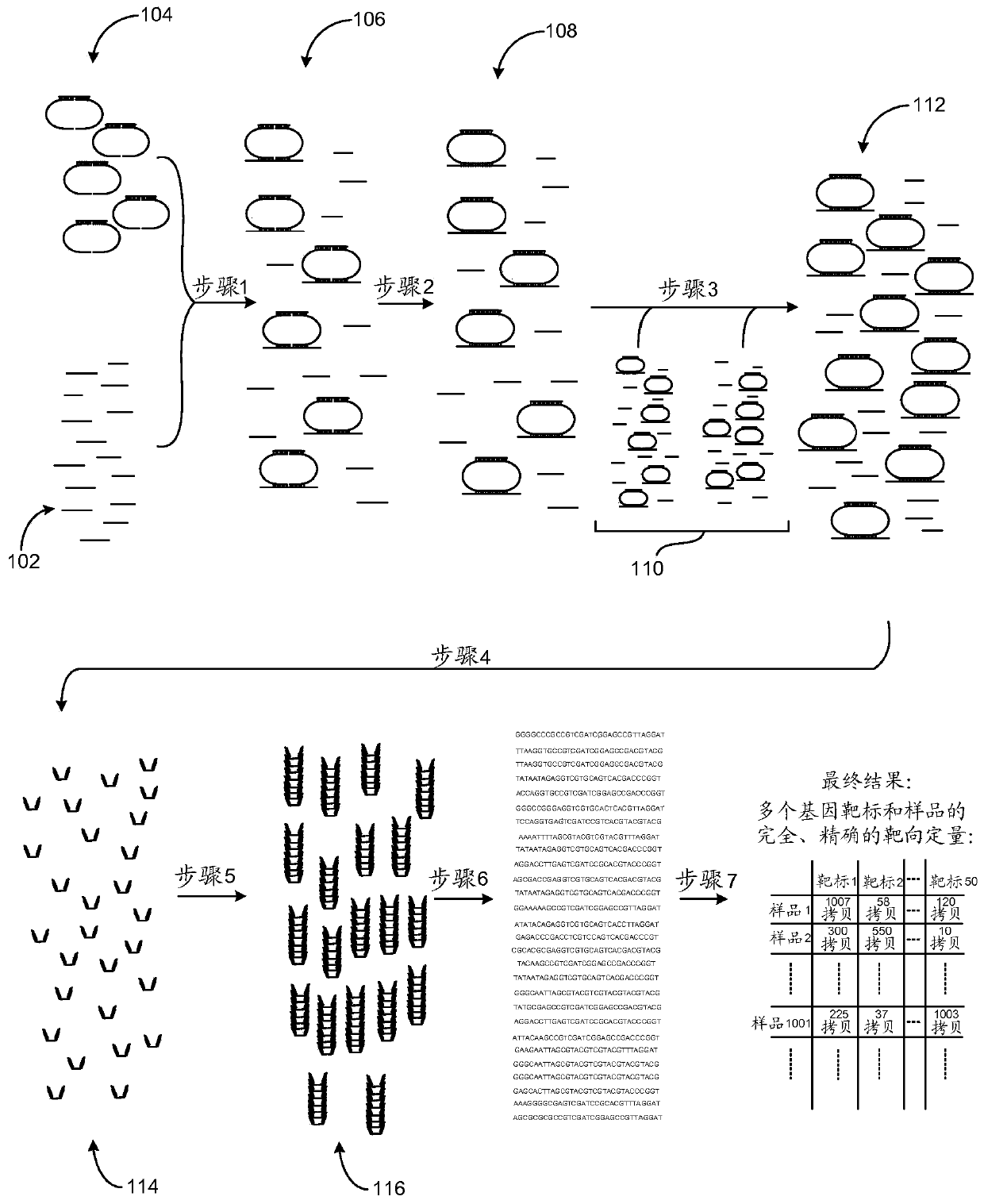
PendingCN111032881AImprove bindingHigh analytical specificityMicrobiological testing/measurementNucleic Acid ProbesDna pooling
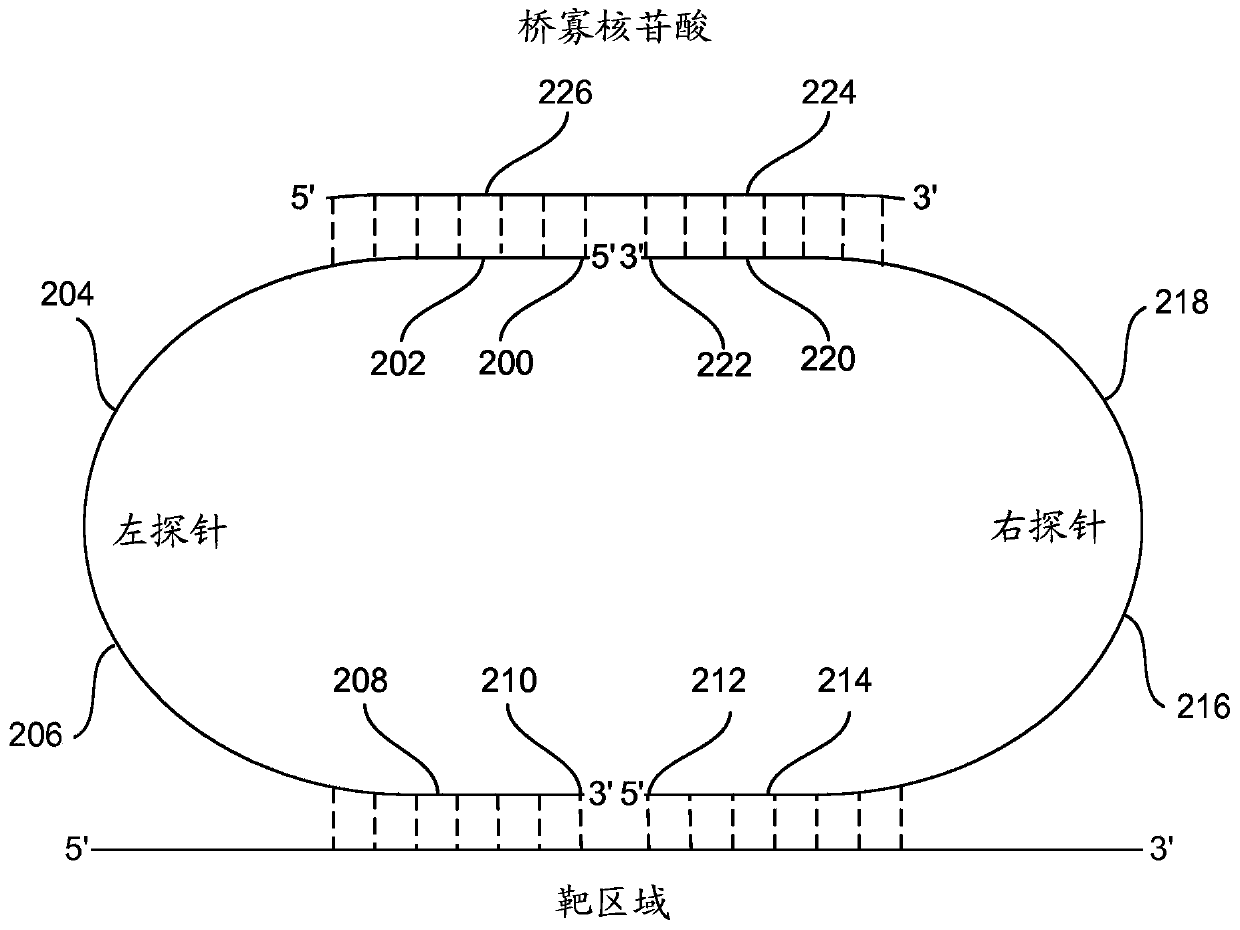
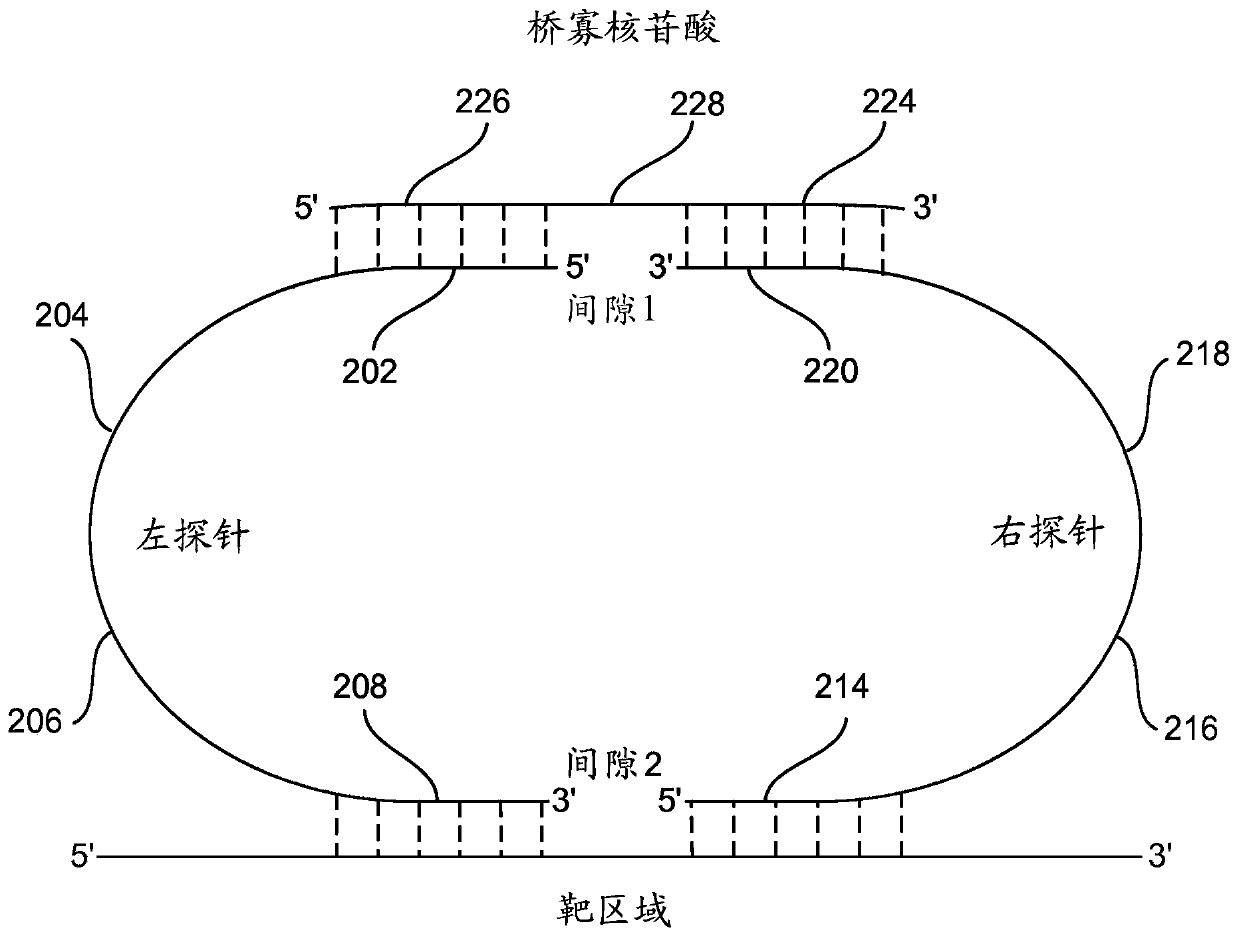
The present invention disclosure relates to a next generation DNA sequencing method and use for accurate and massively parallel quantification of one or more nucleic acid targets. More particularly, the invention is related to the method and a kit comprising probes for detecting and quantifying genetic targets in complex DNA pools primarily used for genetic target and variant detection in human and animal populations and environmental samples. Furthermore, the invention finds particular application in the field of detection of disease-causing genetic alterations in samples obtained from humanbody, including without limiting biopsies, saliva and other secretions, exhaled moisture extracts, tissue, blood plasma (liquid biopsies) or the like. The invention includes one or more target-specific nucleic acid probes per genetic target (left probe and right probe) and a bridge oligo.
Owner:EAWAG SWISS FEDERAL INST OF AQUATIC SCI & TECH
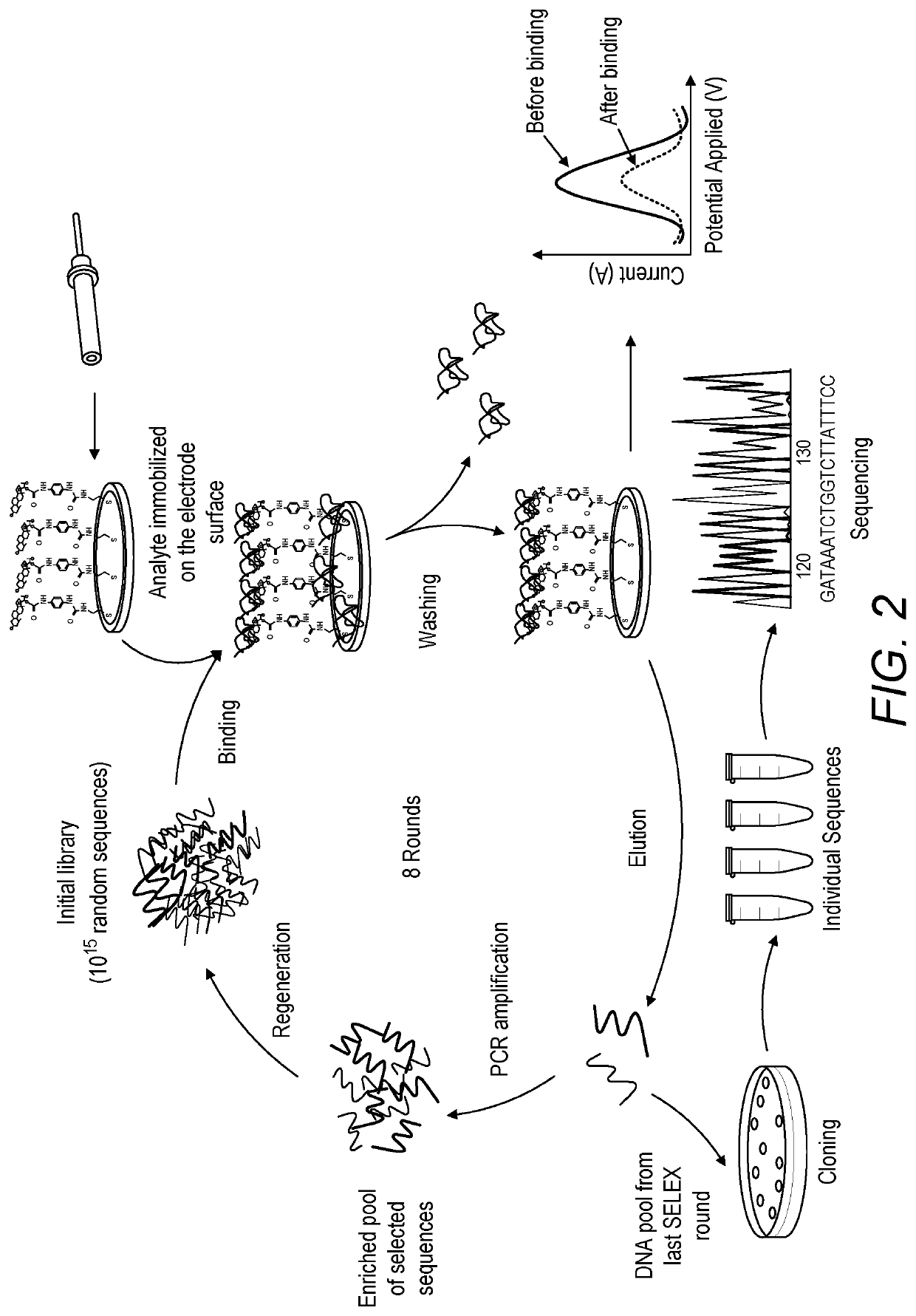
Electrochemical screening for the selection of DNA aptamers
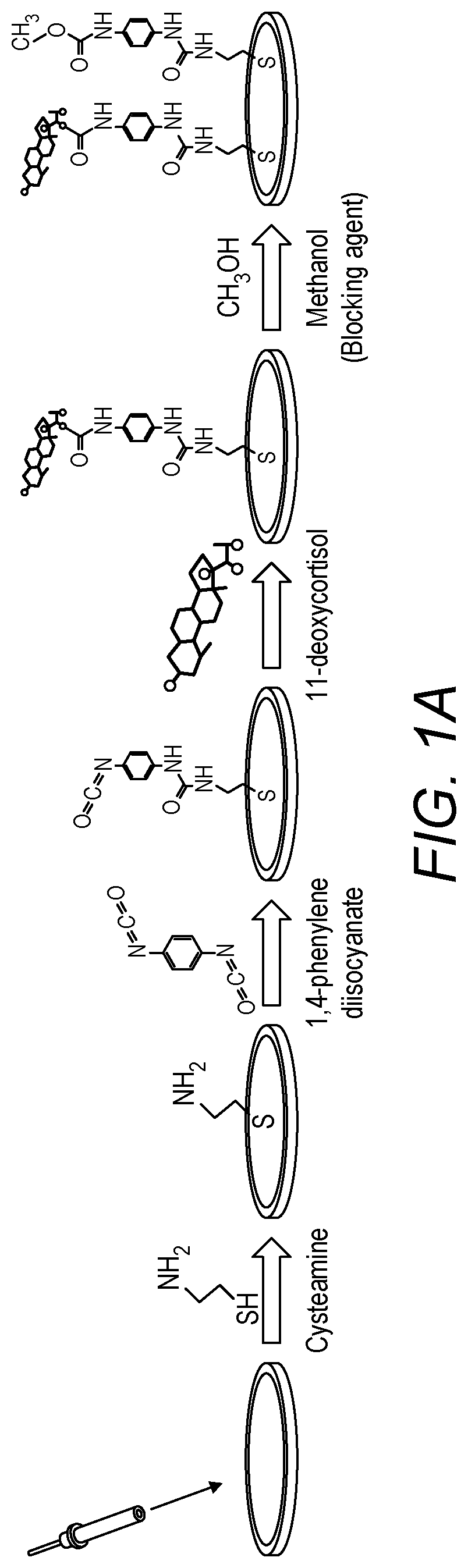
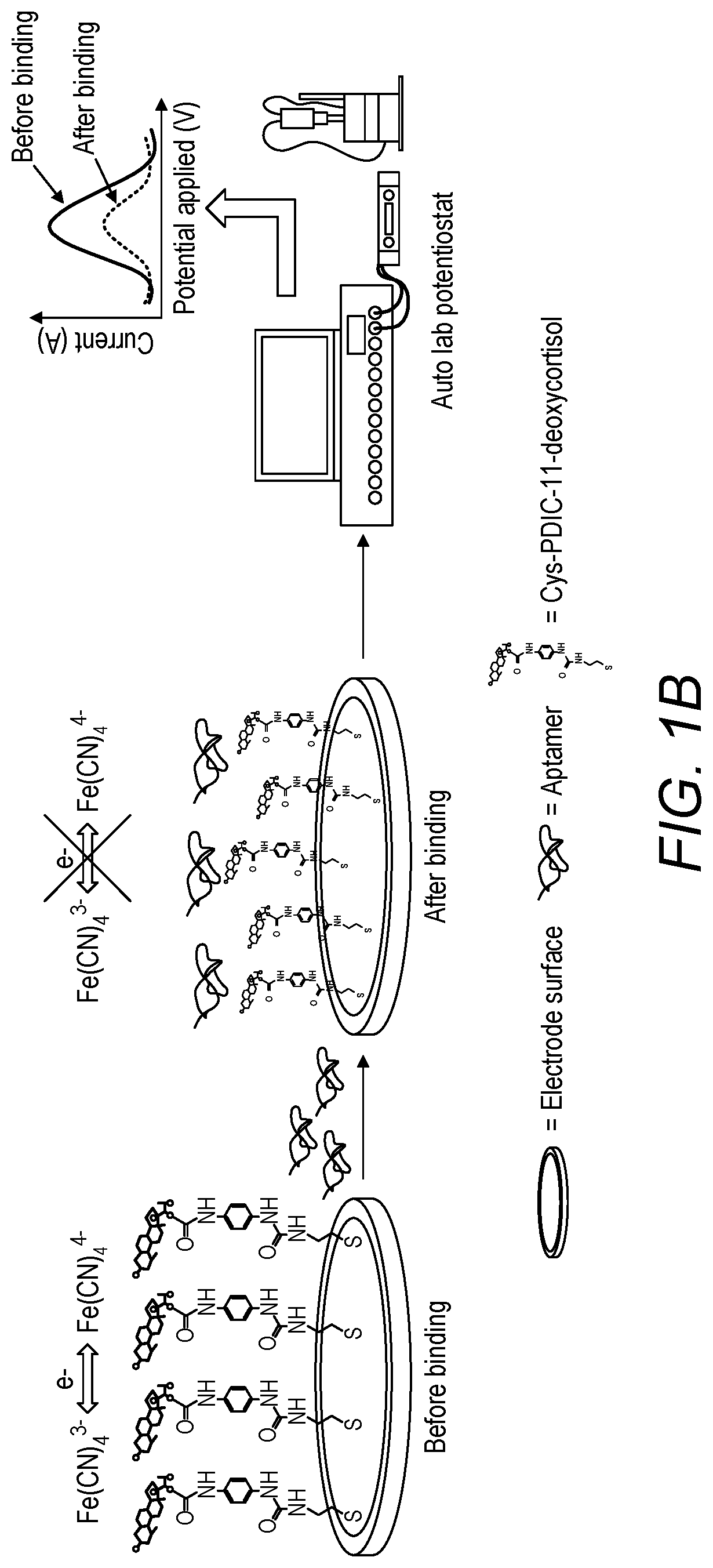
ActiveUS10520462B1Microbiological testing/measurementMaterial analysis by electric/magnetic means11-DesoxycortisolDNA Aptamers
An electrochemical screening method for the selection of DNA aptamers against 11-deoxycortisol (11-DCL) using gold electrode for target immobilization is described. The gold electrode is used as solid matrix instead of the beads for SELEX. The selection steps (SELEX) are performed on the 11-DC modified electrode directly as the DNA library in the first round or the enriched DNA pools in the subsequent rounds were incubated on the electrode, then the unbound DNA is washed and the bound DNA is measured directly by square wave voltammetry. Then elution of the bound DNA is performed for further use.
Owner:ALFAISAL UNIVERSITY
Several Aptamer Sequences Recognizing Benazone and Its Derivatives
ActiveCN106047872BWide range of environmental detection application prospectsRapid detection expansionBiological testingDNA/RNA fragmentationDna poolingSingle strand
The invention provides DNA aptamer sequences for recognizing bentazone and derivatives thereof. DNA aptamer as referred in the invention is single-stranded oligodeoxynucleotide with different oligodeoxynucleotide sequences; and the sequences are capable of recognizing the herbicide bentazone in a solution and applicable as biological recognition materials for detection of the content of bentazone in samples. The DNA aptamer sequences are obtained through in-vitro seletion or SELEX from a random DNA pool. In an aqueous solution, the bonding strength of the DNA aptamer sequences and bentazone molecules dissociation constant enables a dissociation constant (Kd) to reach the magnitude of [mu]M.
Owner:TSINGHUA UNIV
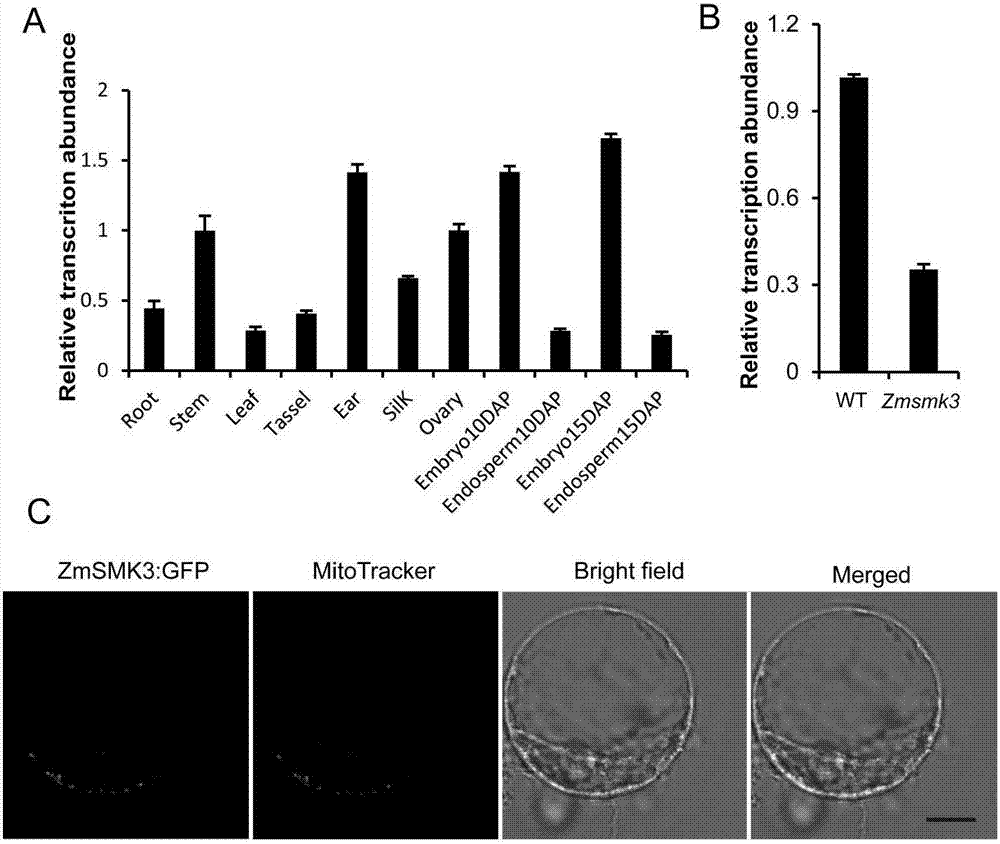
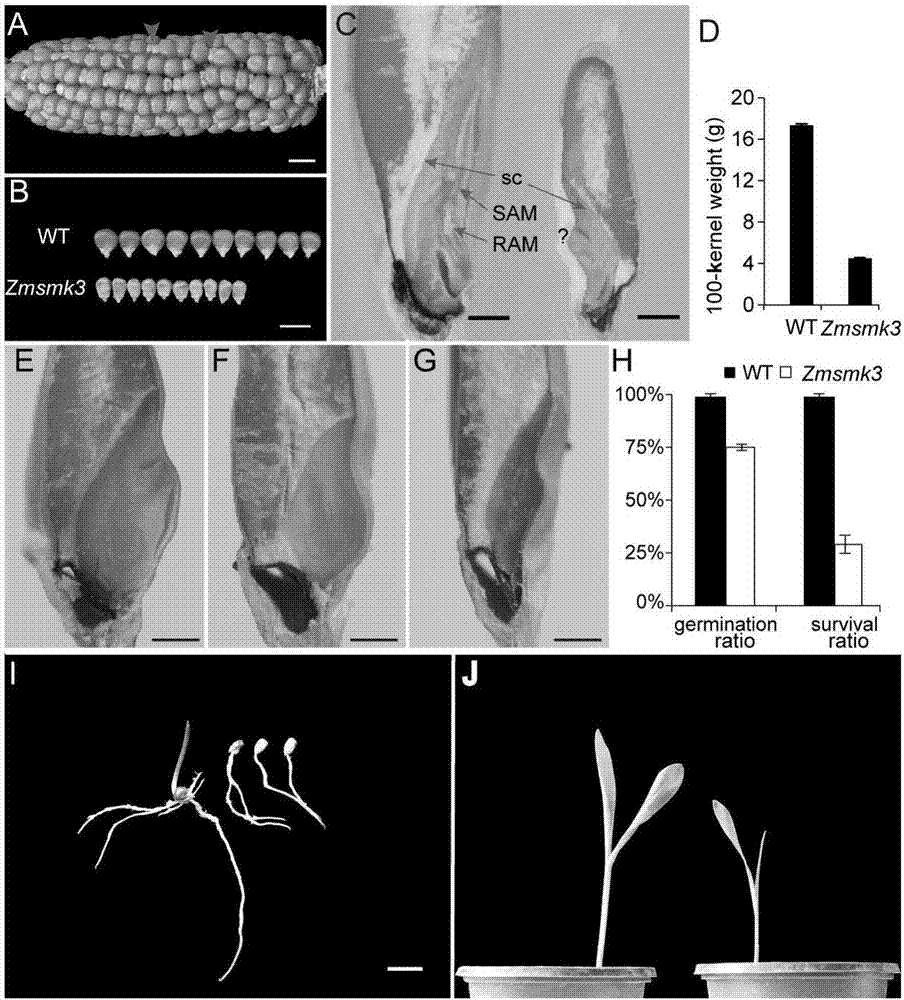
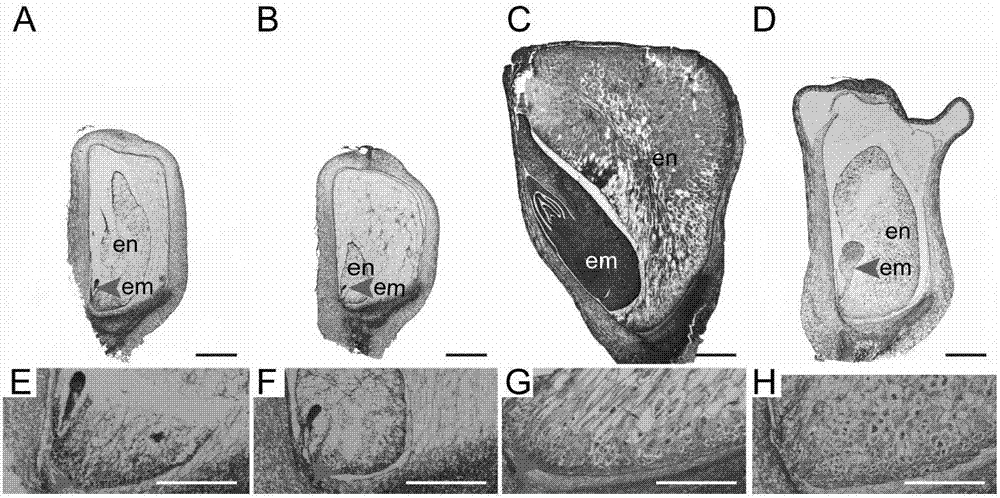
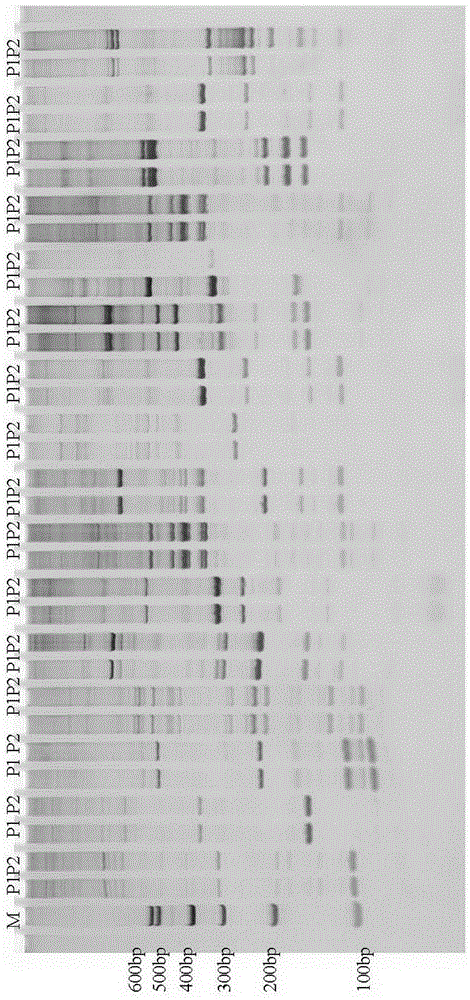
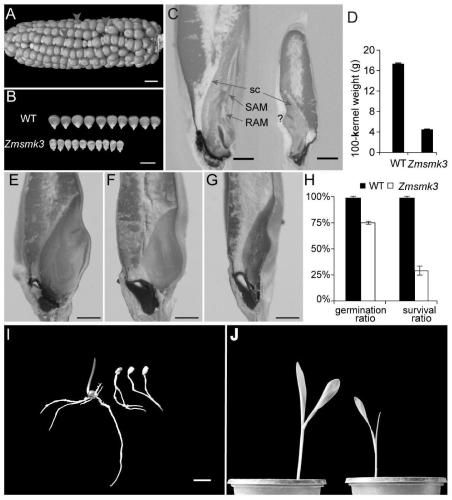
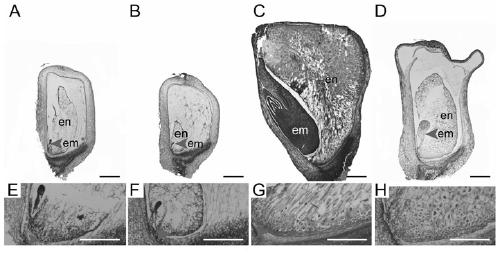
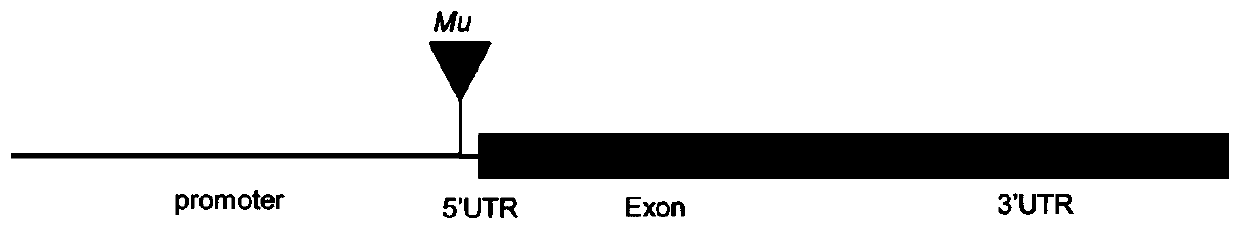
Gene ZmSmk3 for encoding corn mTERF protein, cloning method and application thereof
ActiveCN107475262AAffect assembly efficiencyAffect activityMicrobiological testing/measurementPlant peptidesWild typeDna pooling
The invention relates to a gene ZmSmk3 for encoding corn mTERF protein and a cloning method for the gene ZmSmk3. The cloning method comprises the following steps of: respectively extracting DNA of a plurality of homozygous mutants and homozygous wild type plant leaves, mixing the extracted DNA of the homozygous mutants and constructing a DNA pool of the homozygous mutants; mixing the extracted wild type DNA and constructing a mild type DNA pool; respectively carrying out heat-asymmetric staggering PCR (Polymerase Chain Reaction) on the constructed DNA pool of the homozygous mutants and the wild type DNA pool to obtain PCR product segments; detecting obtained PCR amplification product segments by using agarose gel electrophoresis, recovering specific bands generated in the DNA pool of the homozygous mutants, and carrying out sequencing to obtain a sequence of the mutant gene ZmSmk3. The mutation of the gene ZmSmk3 for encoding the corn mTERF protein has an influence on the seed development progress, and the gene has an important guidance significance for breeding practice.
Owner:HUAZHONG AGRI UNIV
Srap molecular marker closely linked to tomato male sterility gene and its obtaining method
ActiveCN104120126BGuaranteed purityImprove protectionMicrobiological testing/measurementDNA/RNA fragmentationMarker analysisGenotype
The invention discloses an SRAP molecular marker closely linked with male sterility genes of tomatoes and a preparation method thereof. The method comprises the following steps: taking purple-stem fertile tomatoes 87-5 as male parents and green-stem male sterile tomatoes at the seedling stage as female parents, hybridizing to generate F1 generation, and selfing to build an F2 segregation population; performing SRAP marker analysis on male sterility genes ms by adopting a bulked segregant analysis method, randomly combining primers before and after SRAP, selecting 544 pairs of primer combinations, and screening between male sterile and fertile pools to obtain two polymorphic markers C10B9_1 and C10B9_4 between two DNA pools; performing SRAP marker verification on the F2 segregation population by virtue of the two markers, wherein linkage analysis discovers that the linkage distance between the marker C10B9_1 and the sterility genes ms and the linkage distance between the marker C10B9_4 and the sterility genes ms are 3.3cM and -3.3cM respectively. By virtue of the molecular marker, assisted selection of male sterile tomatoes can be performed, the transfer cycle is shortened, the transfer efficiency is improved, a tedious program of identifying the sterility of each generation by selfing in a transfer process is omitted, conventional phenotype selection is transformed into genotype selection, and the accuracy and scientificity of selection are improved.
Owner:HORTICULTURE INST OF XINJIANG ACAD OF AGRI SCI
Method and system for regulating nutrition based on intestinal microorganisms
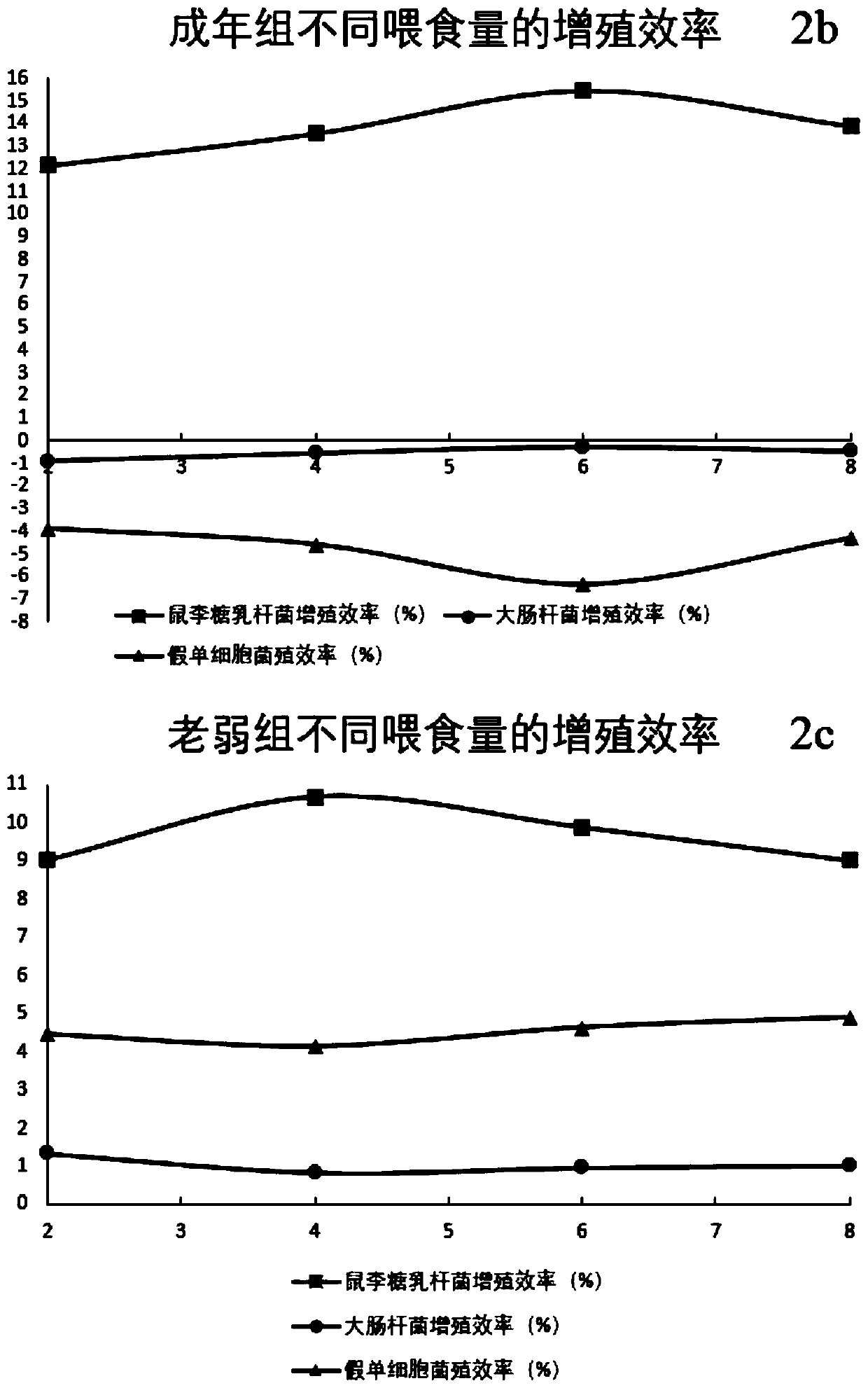
InactiveCN111053244AProliferate fullyStable proliferationFood freezingMicrobiological testing/measurementBiotechnologyFeces
The invention discloses a method for regulating nutrition based on intestinal microorganisms. The method uses nutrition-regulating milk for regulation, and comprises the following steps: S1, selectinga regulation substrate, a regulation flora and a nutrition matrix; S2, preparing a propagation solid-liquid mixture; S3, preparing a nutrition-regulating milk substrate; and S4, preparing the nutrition-regulating milk. The invention further provides a system for regulating nutrition based on intestinal microorganisms. The system comprises the following steps: S1, grouping; S2, feeding in groups,and collecting excrement; S3, constructing a total DNA pool; S4, determining the content of DNA representing bacteria; S5, constructing a nutrition-regulating system model of the feeding amount and the bacterium content. The mutual relationship among the substances of the raw materials is used to enhance the autoimmunity and inhibit the enteropathogenic bacteria, and a feedback regulation mechanism is used to prevent the excessive propagation of flora and regulate the growth of intestinal microorganisms; and the animal model of the nutrition-regulating system is accurately constructed by utilizing a recombinant DNA and polymerase chain reaction technology, so the accuracy of the model is embodied.
Owner:SHANDONG SPORT UNIV
Detection method of Chinese simmental cattle carcass and meat quality trait genetic markers
ActiveCN104059963BSolve the cumbersomeFix instabilityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologySimmental cow
Owner:JILIN UNIV
A gene zmsmk3 encoding maize mterf protein and its cloning method and application
The invention relates to a gene ZmSmk3 for encoding corn mTERF protein and a cloning method for the gene ZmSmk3. The cloning method comprises the following steps of: respectively extracting DNA of a plurality of homozygous mutants and homozygous wild type plant leaves, mixing the extracted DNA of the homozygous mutants and constructing a DNA pool of the homozygous mutants; mixing the extracted wild type DNA and constructing a mild type DNA pool; respectively carrying out heat-asymmetric staggering PCR (Polymerase Chain Reaction) on the constructed DNA pool of the homozygous mutants and the wild type DNA pool to obtain PCR product segments; detecting obtained PCR amplification product segments by using agarose gel electrophoresis, recovering specific bands generated in the DNA pool of the homozygous mutants, and carrying out sequencing to obtain a sequence of the mutant gene ZmSmk3. The mutation of the gene ZmSmk3 for encoding the corn mTERF protein has an influence on the seed development progress, and the gene has an important guidance significance for breeding practice.
Owner:HUAZHONG AGRI UNIV
A traceable SNP molecular marker of porcine sncg gene and its detection method
ActiveCN103589716BMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceIntein
The invention discloses a SNP molecular marker in a porcine SNCG gene for tracing and a detection method thereof. The SNP molecular marker is obtained by DNA pool sequencing and has altogether 1599 bp. The SNP molecular marker contains a part of the sequence of a third exon; a base at a position 984 mutates from 984C to 984T; the DNA sequence of the SNP molecular marker is represented by SEQ ID NO. 1. The results of analysis of the allelic distribution of the SNP molecular marker in a trial group containing 11 pig species / strains show that allelic frequencies of the SNP molecular marker in different species or strains are approximate, polymorphism is rich, allelic frequency distribution difference between species or strains is small, the degree of heterozygosis is all larger than 0.3, and thus, the SNP molecular marker disclosed herein can be used for DNA tracing for pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
A method for gene mapping of Brassica napus wax powder based on whole genome resequencing
ActiveCN111575399BSequencing data is sufficientQuality standardsMicrobiological testing/measurementSequence analysisBiotechnologyGenome resequencing
The invention provides a gene positioning method of brassica napus wax powder based on whole genome resequencing, which belongs to the technical field of biological information. The present invention establishes near-isogenic lines on the basis of segregating populations. Using mixed grouping analysis, non-wax plants in the 3:1 segregation population of near isogenic lines, all non-segregation populations with wax phenotype and 3 parents were selected, and 5 DNA pools were constructed for genome resequencing. The resequencing data was used for genetic association analysis, and the loci controlling the traits of wax powder were located in the 590663-1657546bp region of the A08 chromosome. The biggest feature of the method of the present invention is that it does not need to detect all progeny individuals, but conducts mixed pool analysis on the progeny of two extreme traits. Moreover, the sequencing data of the method of the present invention are sufficient and of acceptable quality, and can be used for the next step of analysis.
Owner:SHANGHAI ACAD OF AGRI SCI
A method for detecting beef cattle plag1 gene indel marker and its special kit
InactiveCN107988385BImprove accuracyImprove stabilityMicrobiological testing/measurementDNA/RNA fragmentationDna poolingZoology
The invention discloses a method for detecting Indel markers of beef cattle PLAG1 genes and a special kit thereof. The method comprises the following steps: based on a PCR (Polymerase Chain Reaction)technology, by taking a blood genome DNA pool of beef cattle (Jiaxian red cattle) as a template, and amplifying Indel sites of cattle PLAG1 genes by utilizing a specific primer pair P1; and performingagarose gel electrophoresis, and dividing different individuals into a deletion form, a normal form and a hybrid form according to electrophoresis results. The method provided by the invention is established on a relevance basis between the Indel sites of beef cattle PLAG1 genes and growth traits, and the method is simple, rapid and convenient to popularize and use and contributes to acceleratingmolecular marker-assisted selection of the beef cattle.
Owner:NORTHWEST A & F UNIV
A Traceable SNP Molecular Marker of Pig Amy2 Gene and Its Detection Method
ActiveCN103589715BMicrobiological testing/measurementAnimals/human peptidesAgricultural scienceIntein
The invention discloses an SNP molecular marker in a porcine AMY2 gene for tracing and a detection method thereof. The SNP molecular marker is obtained through DNA pool sequencing and has altogether 1060 bp. The SNP molecular marker contains a part of the sequence of a fifth exon, a part of the sequence of a seventh exon and the complete sequences of a fifth intron, a sixth exon and a sixth intron; a base at a position 104 mutates from 104 C to 104 T; the DNA sequence of the SNP molecular marker is represented by SEQ ID NO. 1, and an amino acid sequence encoded by the SNP molecular marker is represented by SEQ ID NO. 2. The SNP molecular marker is used in cultivation of commercial pigs and is extensively applicable to DNA tracing and detection for pork products, especially to security tracing for pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
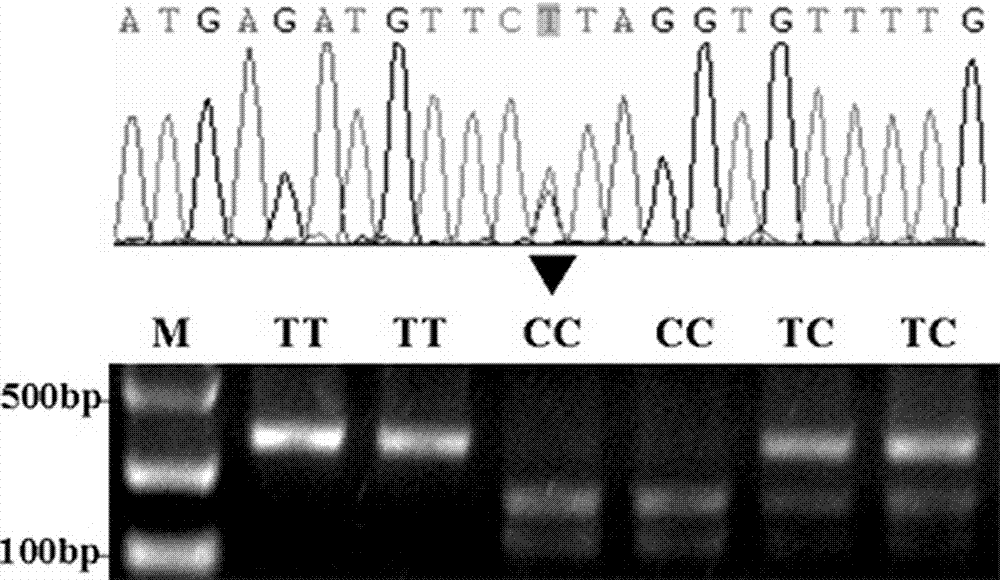
Method for detecting single nucleotide polymorphism of sheep DAQL gene by using PCR-RFLP and application of method
ActiveCN110592190AImprove accuracySave genotyping stepsFood processingMicrobiological testing/measurementStatistical analysisMathematical model
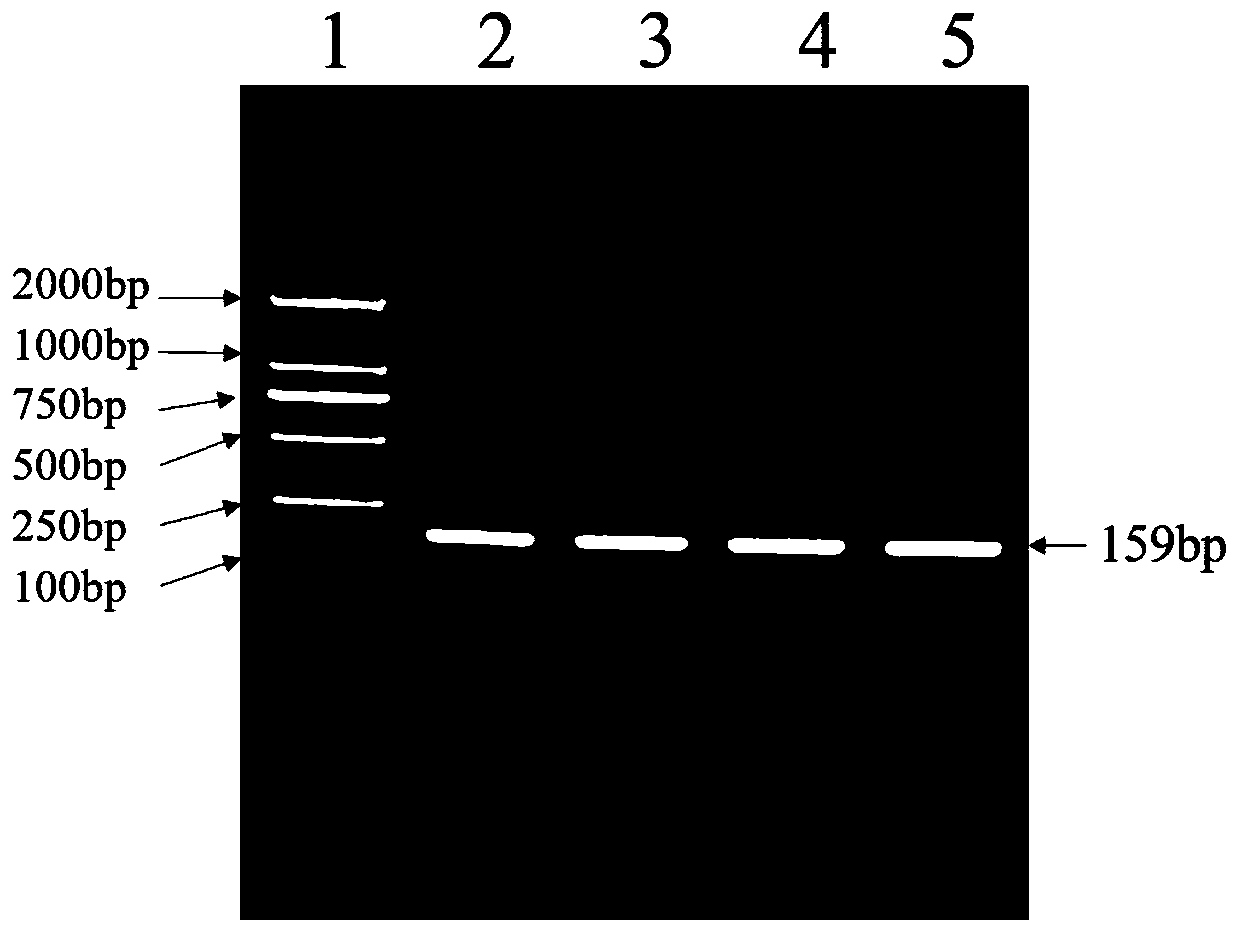
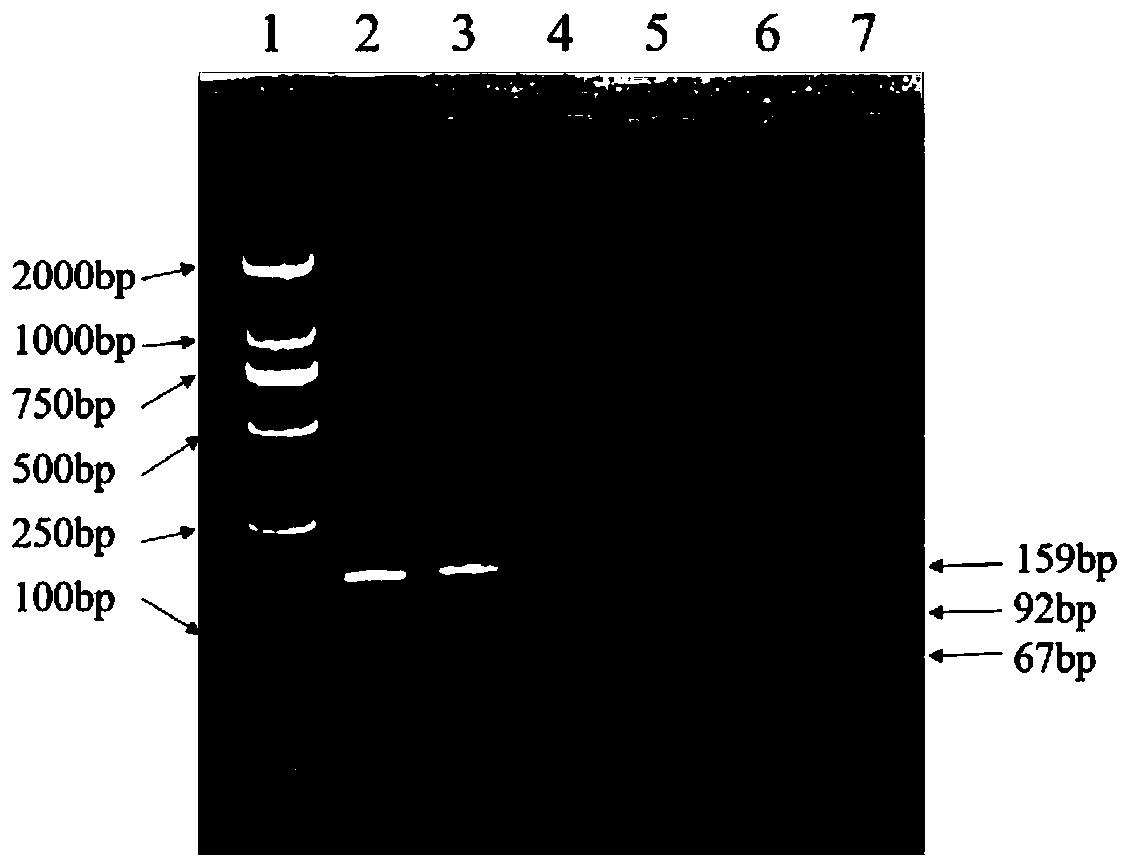
The invention discloses a method for detecting a single nucleotide polymorphism of sheep DAQL gene by using PCR-RFLP, and particularly relates to a method for detecting a single nucleotide polymorphism of sheep DAQL gene with amino acid at 26750th position being G or A by using PCR-RFLP. The method comprises the following steps: S1, collecting a sample and performing early-stage treatment on the sample; S2, constructing a variety DNA pool; S3, treating the sample by a PCR-RFLP method; and S4, gel imaging to determine genotype; and two-way sequencing to determine the single nucleotide polymorphism. The invention also provides an application of the method for detecting single nucleotide polymorphism of sheep DAQL gene by using PCR-RFLP. The application comprises the following steps: I analysis of phenotypic data and statistical analysis of single nucleotide loci; II establishing a mathematical model through association analysis; and III constructing a model to derive the single nucleotide polymorphism of the DAQL gene in adult sheep. The method and the application have beneficial effects that: the variety DNA pool is treated by PCR-RFLP to achieve a purpose of accurate labeling; digestion with AluI enzyme is used to solve a problem of false positives; and the mathematical model is used for correlation analysis to improve accuracy of molecular markers.
Owner:甘肃润牧生物工程有限责任公司
Breeding method of high-yielding Jersey cows
ActiveCN108823320BImprove breeding efficiencyRapid Breeding of CattleMicrobiological testing/measurementMilk cow'sDna pooling
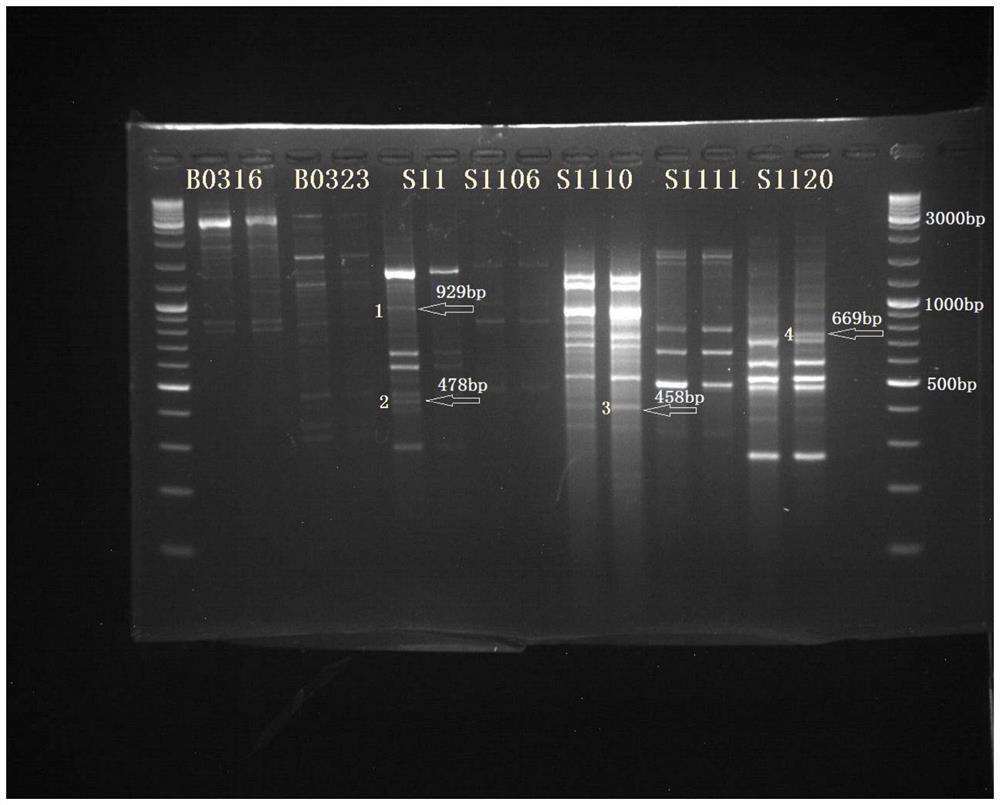
The invention belongs to the technical field of bioengineering, in particular to a breeding method for high-yielding Jersey cows. In this application, 250 10-base random primers were used to perform RAPD-PCR between two DNA pools composed of 20 high-yield (305d milk yield>4000kg) and 20 low-yield (305d milk yield<3200kg) Jersey cattle. Amplified molecular marker screening method. All individuals were tested by PCR with 200 primers screened, and three RAPD markers related to milk production traits of dairy cows were obtained, and they were cloned, sequenced and analyzed by bioinformatics. The test results showed that the DNA fragments of S11, S1110 and S1120 were related to milk yield, among which S11 was positively correlated with milk yield traits, and S1110 and S1120 were negatively correlated with milk yield traits.
Owner:GUANGXI ZHUANG AUTONOMOUS REGION INST OF ANIMAL HUSBANDRY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com