Accurate and massively parallel quantification of nucleic acid
An oligonucleotide and labeling nucleotide technology, applied in the field of a method and a kit containing probes, can solve the problem of high cost of sequencing library preparation, waste of unrelated locus sequencing work, limited interpretation data statistical selection, etc. problem, to achieve high analytical specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0039] The following detailed description illustrates embodiments of the invention and how to achieve it. Although a few modes for carrying out the invention have been disclosed, those skilled in the art will recognize that other embodiments for carrying out or practicing the invention are also possible.
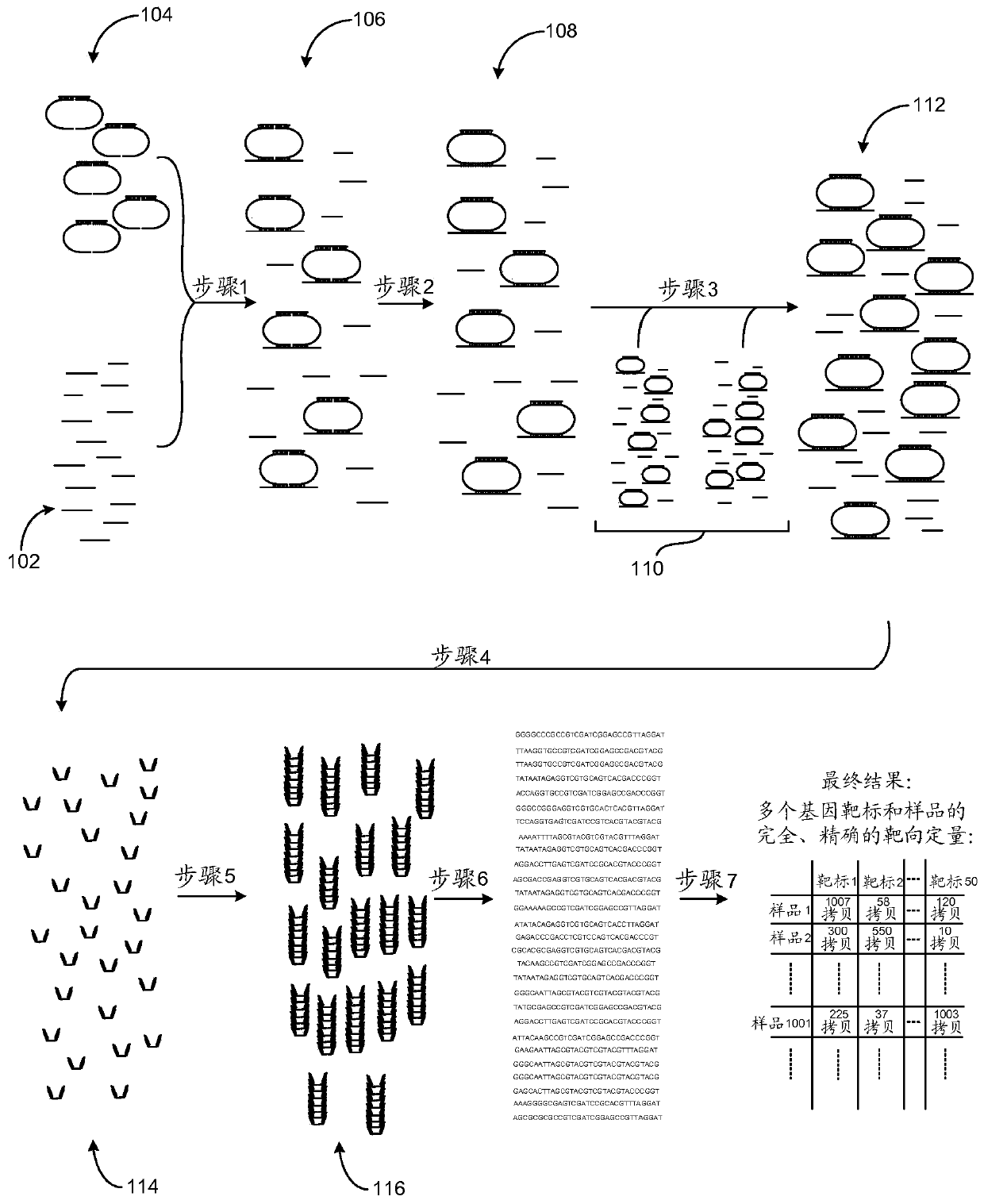
[0040] In a first embodiment, the present invention provides a method for high-throughput detection of one or more target nucleotide sequences in a plurality of samples, the method comprising the steps of:
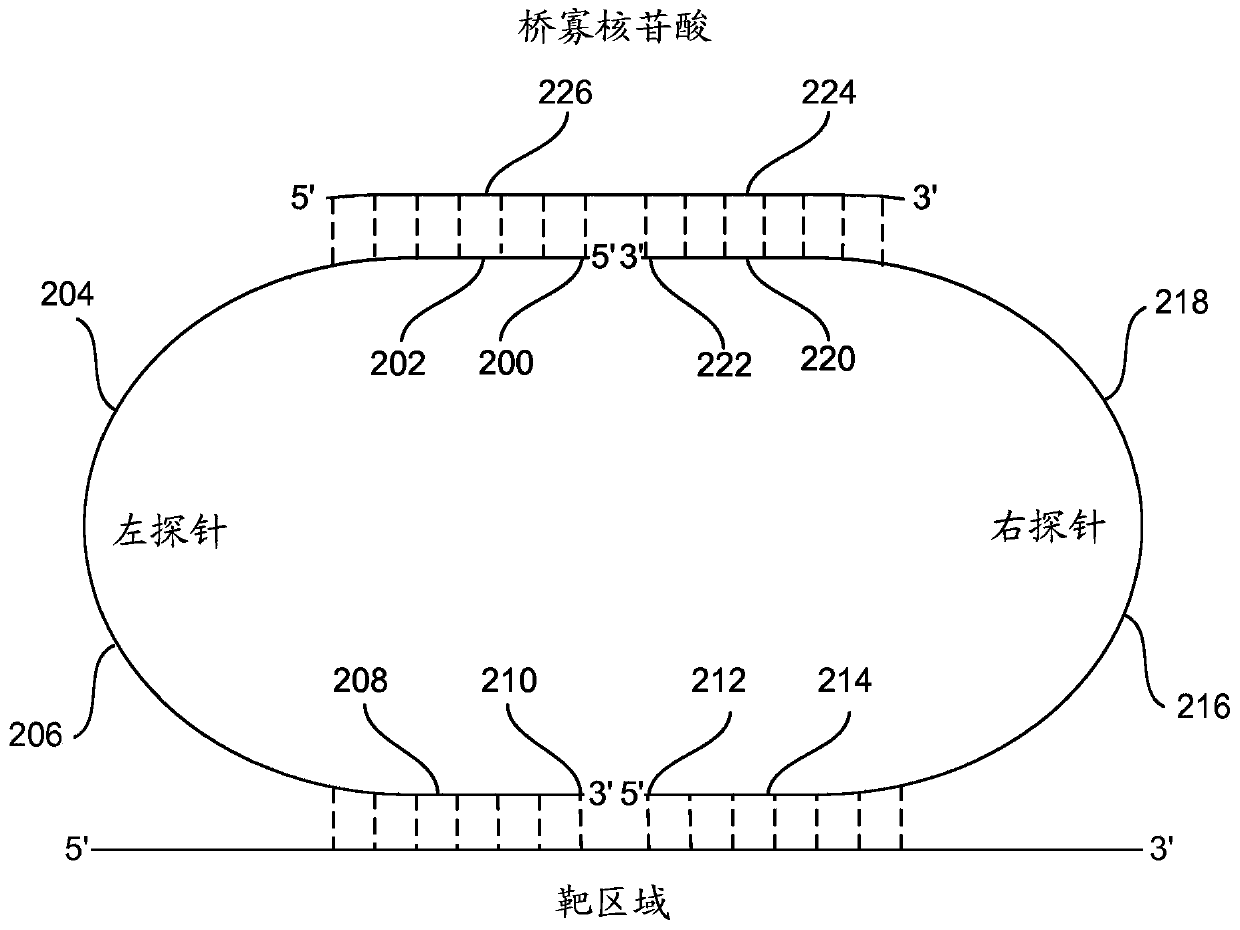
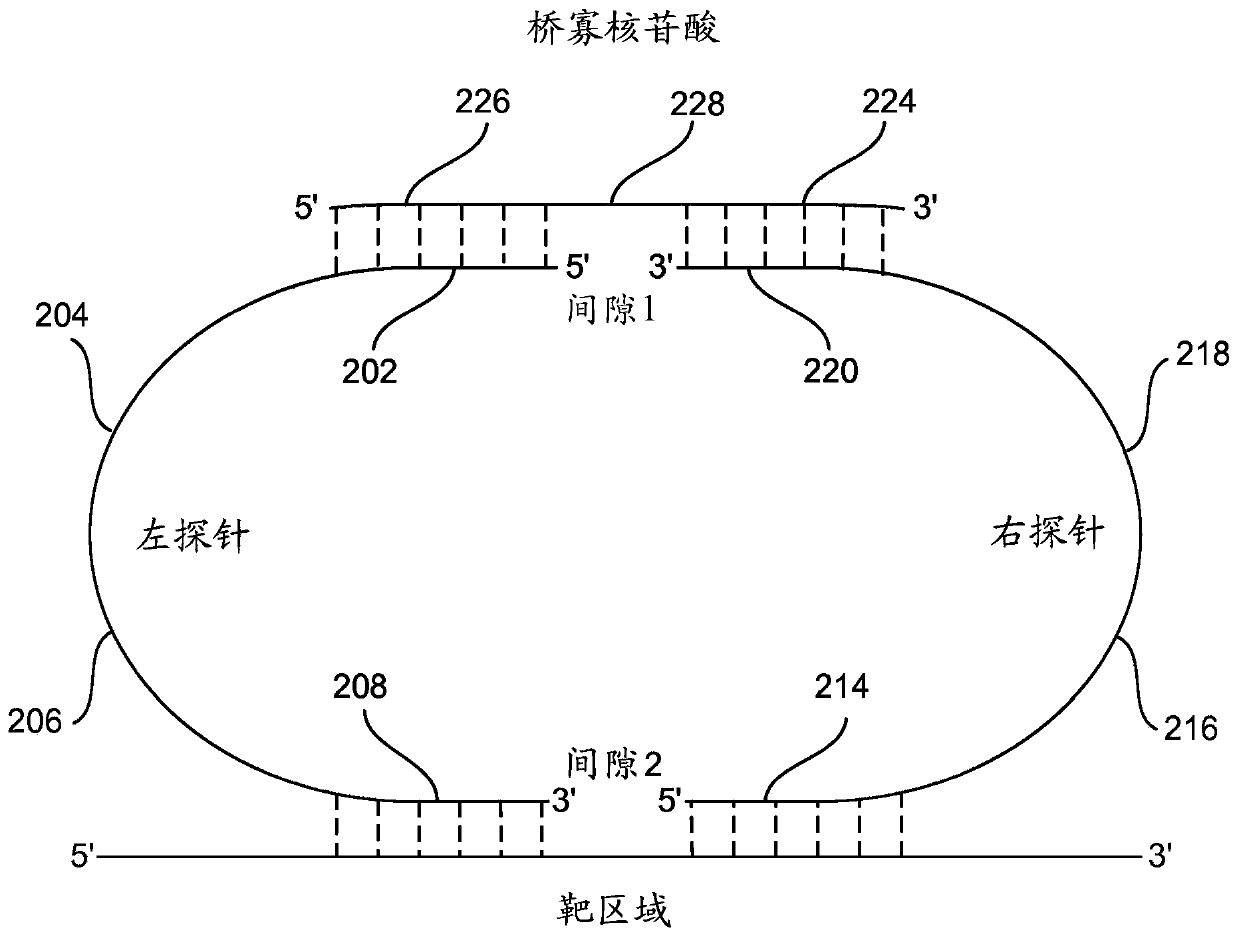
[0041] (i) providing for each target nucleotide sequence in each sample: a first probe, a second probe, and a bridge oligonucleotide, wherein
[0042] The first probe includes an optional 5' phosphate starting at the 5' end of the molecule, a first bridge oligonucleotide specific sequence, an optional first universal sequence, an optional first sequence barcode, and a first target-specific portion at the 3' end of the probe; and wherein
[0043] The second probe includ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com