Gene ZmSmk3 for encoding corn mTERF protein, cloning method and application thereof
A cloning method, corn technology, applied in the fields of application, genetic engineering, plant gene improvement, etc., can solve the problems that the function of mTERF protein has not yet been resolved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Cloning of embodiment 1 ZmSmk3 mutant gene
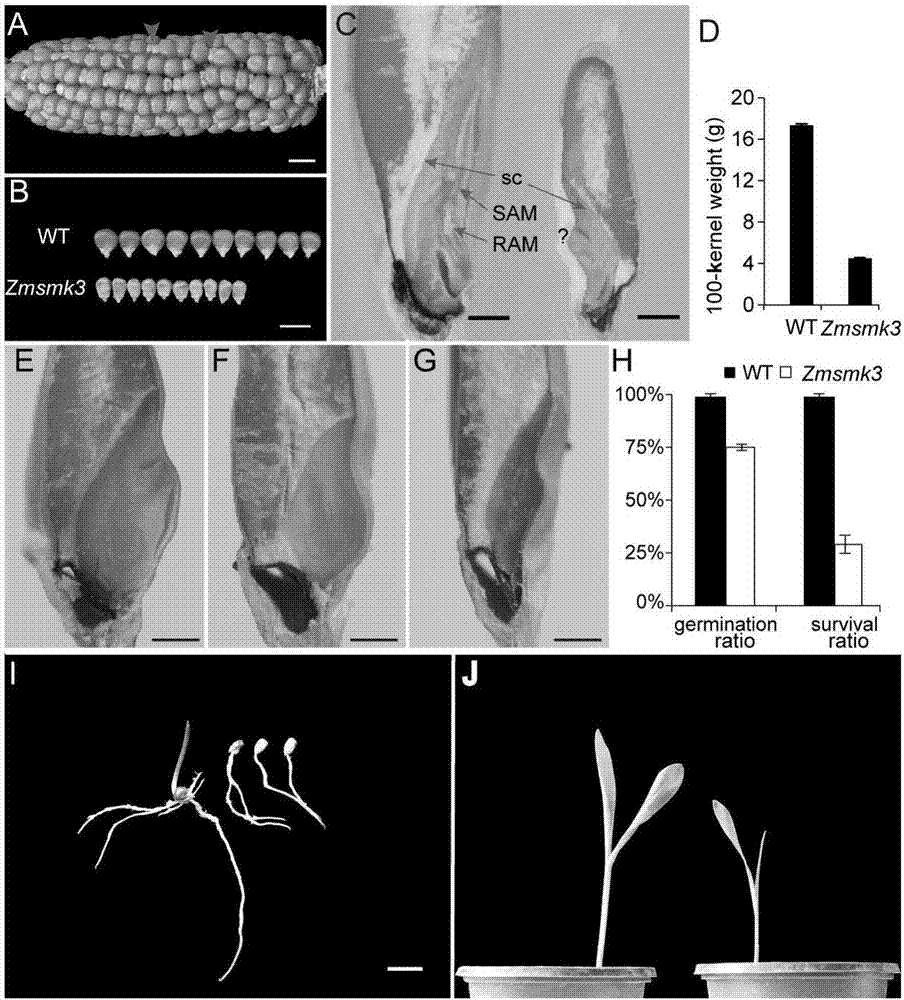
[0066] There were 1 / 4 small-grain mutations in ZmSmk3 heterozygous selfed ears, and the wild type:mutant match was 3:1 (WT:Zmsmk3=1651:526, p=0.37), which proved that the mutant trait was controlled by a recessive single gene. The leaf DNA of 10 homozygous mutants (number: RS1-RS10) and 5 homozygous wild-type plants (number: RW1-5) were extracted by CTAB method. After the concentration was determined, all of them were diluted to 50ng / ul. 5. RS4-8 and RS6-10 were mixed in equal amounts to construct 3 homozygous mutant DNA pools; RW1-5 were mixed in equal amounts to construct a wild-type DNA pool, and the four mixed pools were respectively used as templates for subsequent PCR reactions.
[0067] Use thermal asymmetric staggered PCR, that is, use specific primers TIR9-1, TIR9-2 and random primers for the mu transposon end sequence in Table 5 to amplify. For specific amplification methods, refer to: Liu Wenting, 2006; Settles et ...
Embodiment 2
[0070] Example 2 Detection of mutant gene Zmsmk3 in individual maize plants
[0071] The sequence of the mutant gene Zmsmk3 was compared with the maize genome to find out the genomic position corresponding to the specific fragment. Centering on the insertion site of the matching position, design primers in combination with TIR6 to detect the genotype of a single plant. If the genotype and phenotype are co-segregated, the gene inserted at this site is the candidate gene, further expanding the population verification. The primer used in this paper to identify co-segregation is S21F / R. Its nucleotide sequence:
[0072] S21F: GGAGGAGCTGGTGTAGATCG
[0073] S21R:TACGTGCCAGATTTGATGCT
[0074] TIR6AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC
[0075] Genotype detection uses S21F / R, S21F+TIR6, S21R+TIR6, three pairs of primers to detect the genotype of each individual plant. If only S21F / R has a corresponding PCR product in the amplified bands of the three pairs of primers in a certain indiv...
Embodiment 3
[0090] Example 3 Expression Analysis and Subcellular Localization of ZmSmk3 Gene
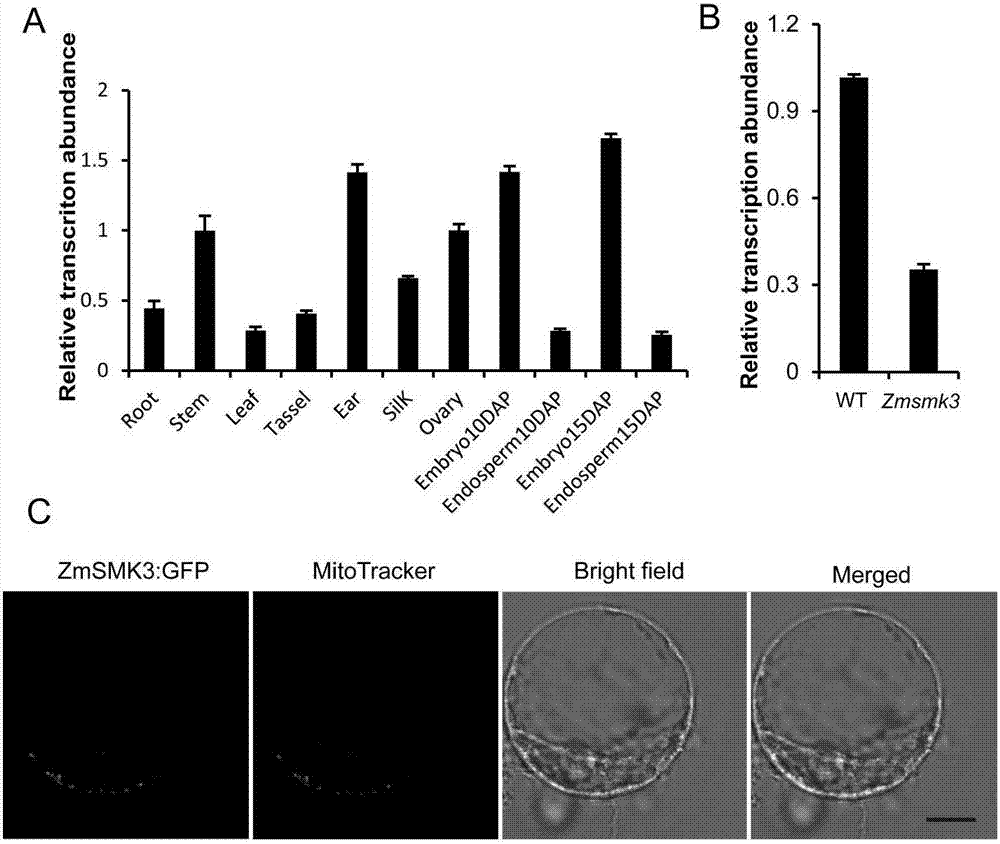
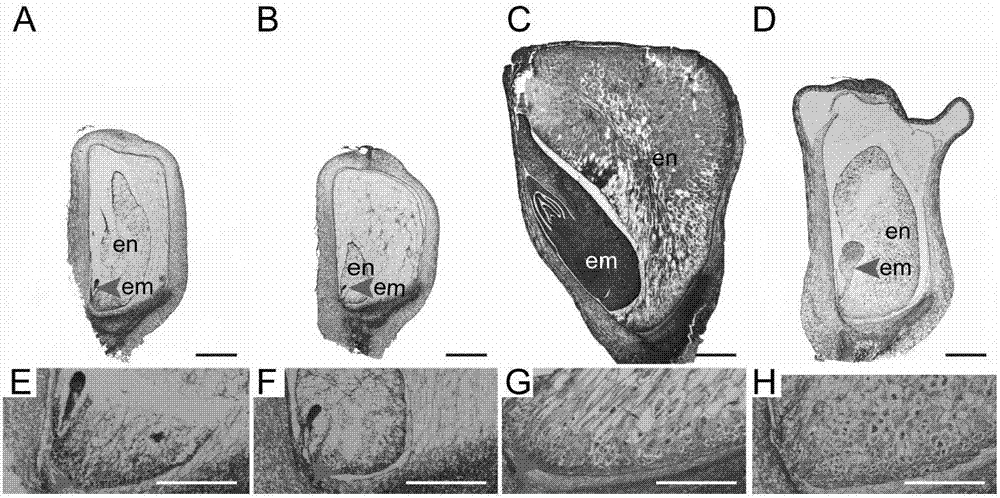
[0091] The expression of ZmSmk3 gene in maize tissues was analyzed by qRT-PCR. ZmSmk3 is a constitutively expressed gene. ZmSmk3 gene is expressed in all vegetative and sexual reproductive tissues, and the expression level is relatively high in the stem, ear, ovary and embryo; the expression level is low in the root , leaves, tassels, filaments and endosperm, results such as Figure 5 Shown in A: qRT-PCR detection of the expression of ZmSmk3 in roots, stems, leaves, tassels, ears, filaments, ovaries, and embryos and endosperms of 15DAP. This result indicates that the ZmSmk3 gene is constitutively expressed in various stages of plant growth and development and in various tissues. At the same time, in the seeds 15 days after pollination, it was detected that the expression level of ZmSmk3 gene decreased in mutants (results such as Figure 5 B).
[0092] The gene was connected to the subcellula...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com