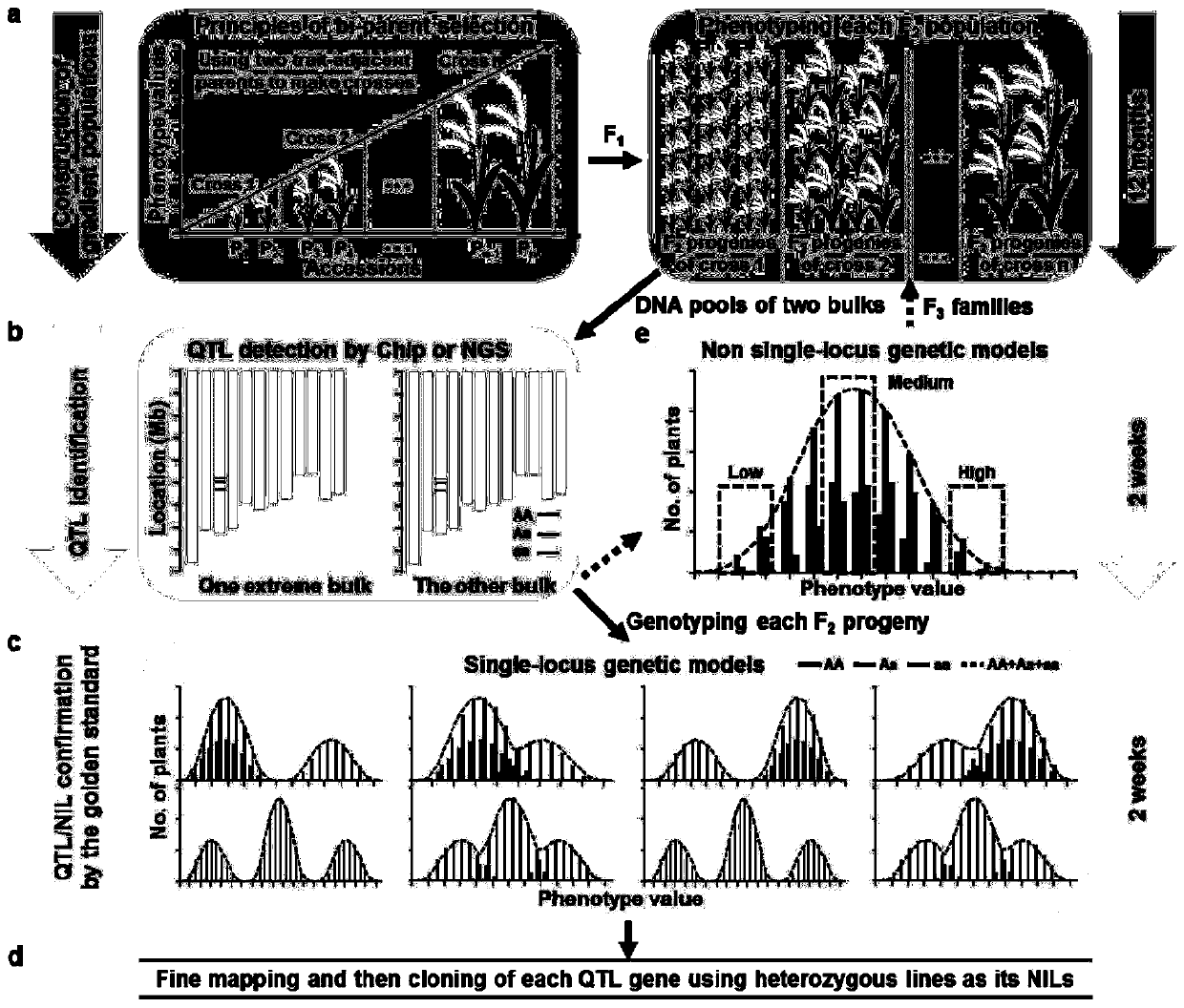
RapMap method for rapid and high-throughput positioning and cloning of plant QTL gene
A high-throughput and fast technology, applied in genomics, bioinformatics, instruments, etc., can solve problems such as low efficacy and slow decay of local linkage disequilibrium, and achieve the goal of overcoming shortcomings and limitations, speeding up speed, and solving complexity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
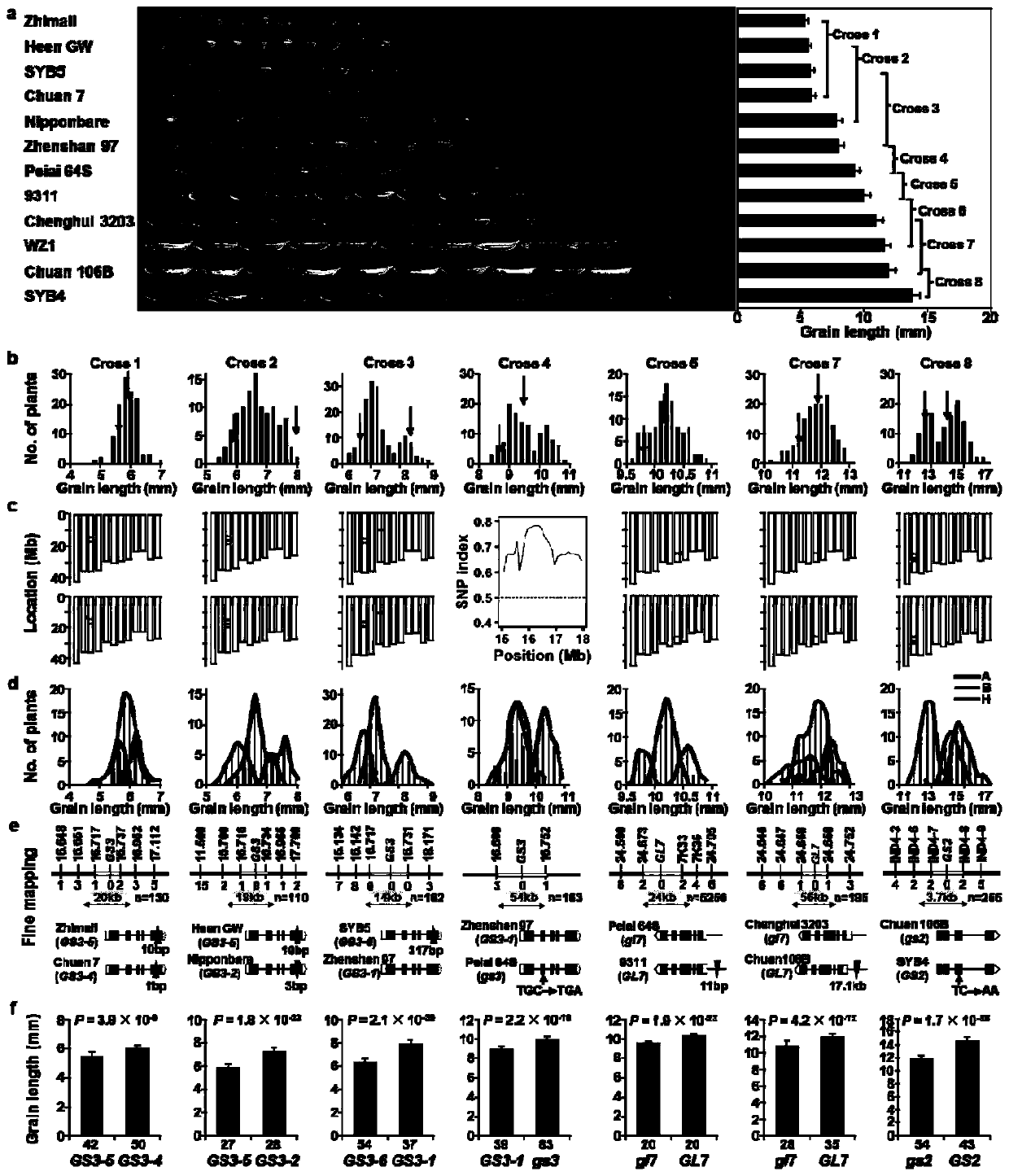
[0032] Embodiment 1: The present invention is applied to the cloning of the QTL gene of rice grain length natural variation.
[0033] We selected 12 parental materials with representative grain length phenotypes ( figure 2 a), in Hainan in 2016, we prepared 8 hybrid combinations with parents with similar grain size, and the F of each combination 1 The seeds were propagated in Wuhan in 2016, and about 200 offspring and their genomic DNA were obtained from each gradient population in Hainan in 2017. Harvest F 2 After the seeds, in 2017, Wuhan carried out the test of grain length phenotype, and constructed a mixed pool with high value (last 15%) and low value (top 15%) ( figure 2 b). Mixed pools of 6 gradient populations (populations 1, 2, 3, 5, 7, 8) were genotyped and QTL mapped using the RICE6K commercial chip (China Seed Group Corporation), which was designed based on the above microcore collection , including four million SNP loci (Yu et al., 2014), the SNP detected by...
Embodiment 2
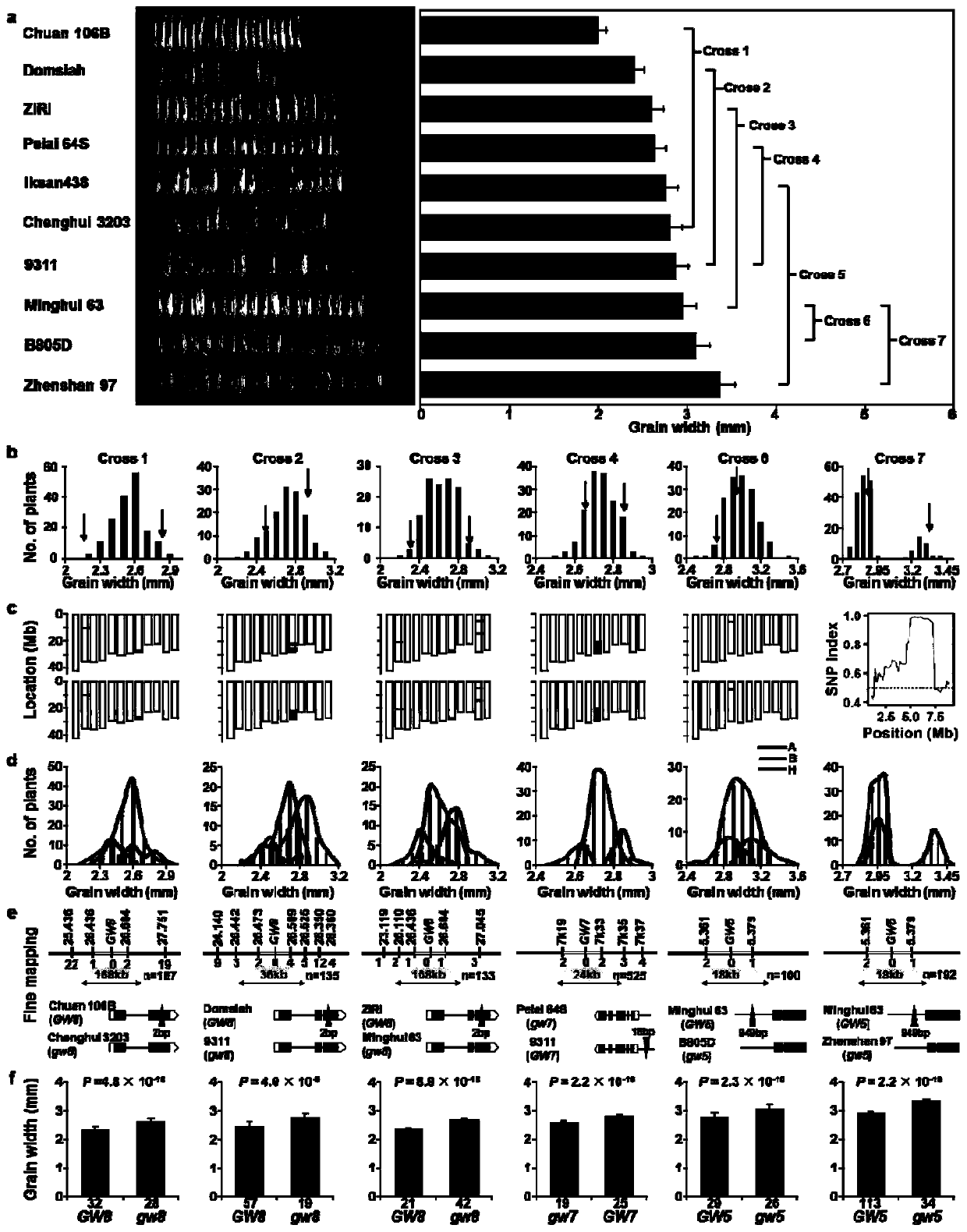
[0035] Embodiment 2: The present invention is applied to the cloning of QTL genes for natural variation of rice grain width.
[0036] While cloning the grain length gene, we selected 10 parental materials with representative grain width phenotypes from the above-mentioned rice micro-core germplasm (grain width variation range 1.99mm-3.43mm) ( image 3 a), In March 2016, 7 hybrid combinations were made in Hainan with parents with similar grain width. It should be noted here that as long as the difference in grain width between the two parents of the hybrid combination is not too large, it is suitable for RapMap, that is, the segregation population controlled by a single locus can be obtained as much as possible and a suitable near-isogenic line can be screened. f 1 The seeds were propagated in Wuhan in 2016 and planted in Jiadai, Hainan in 2017, and each group harvested 200 F 2 The seeds of a single plant were tested in Wuhan in 2017 for the grain width phenotype, and a singl...
Embodiment 3
[0044] The six genes cloned in the present invention contribute greatly to the inheritance of grain length and grain shape in the rice world micro-core collection.
[0045] Utilize the functional variation of 6 control grain length and grain width genes cloned in Example 1 and Example 2 in the genotype of the micro-core collection and the grain length, grain width, aspect ratio, thousand-grain weight, and grain number of the micro-core collection Multiple linear regression analysis was performed on the phenotypes of the six traits of yield per plant to evaluate the genetic contributions of these genes to the six traits, and it was found that three grain length genes could explain 49% of the grain length variation ( Figure 4 a), three grain width genes can explain 53% of the grain width variation ( Figure 4 c), 6 genes can explain 64%, 17%, 15%, and 13% of the phenotypic variation in aspect ratio, thousand-grain weight, grain number, and yield per plant, respectively ( Figu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com