Brassica napus wax powder gene positioning method based on whole genome resequencing
A Brassica napus, the whole genome technology, applied in the field of bioinformatics, can solve the problems of low detection efficiency and heavy workload, and achieve the effect of qualified quality and sufficient sequencing data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
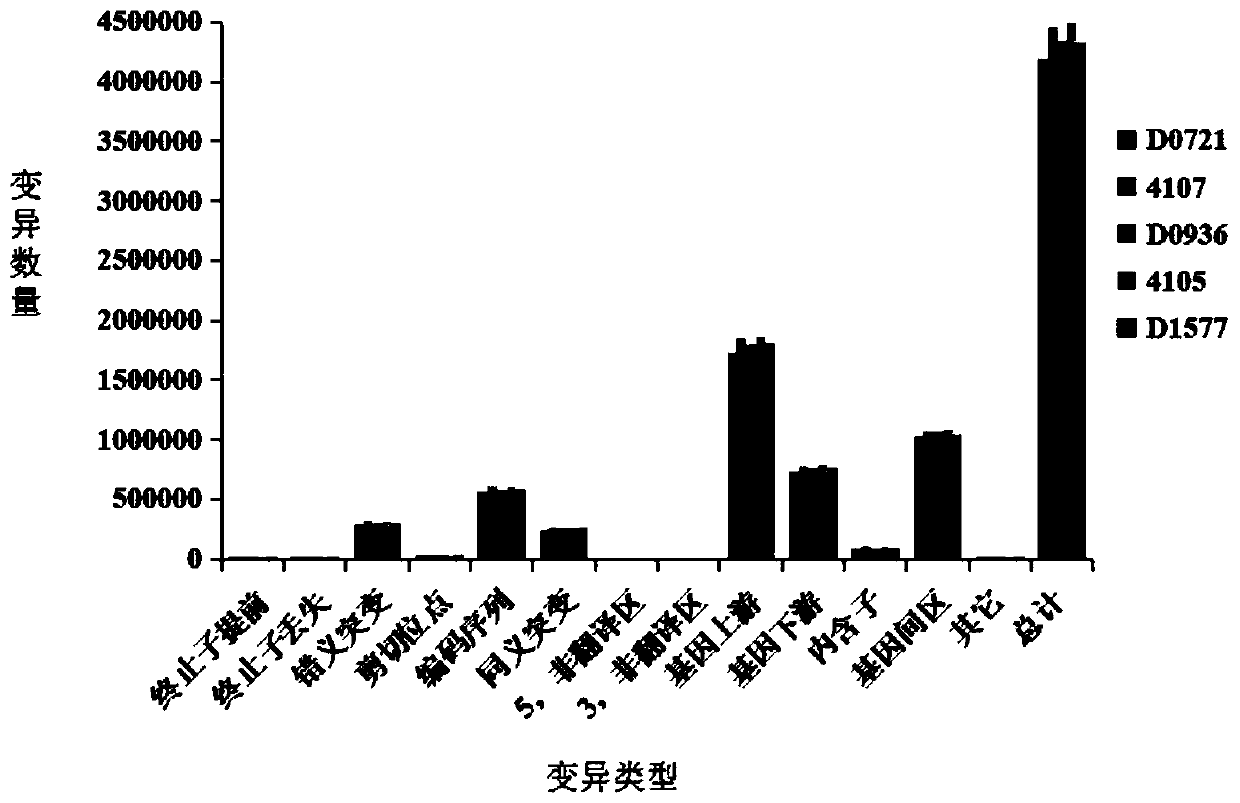
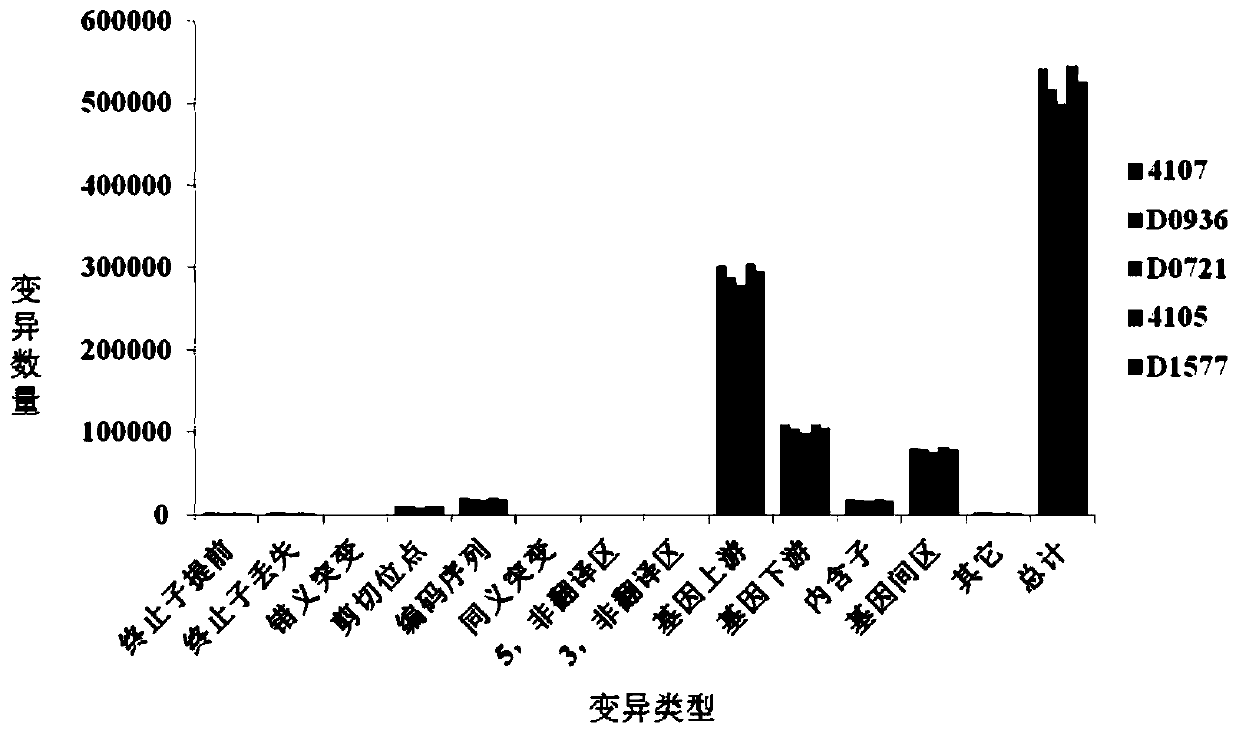
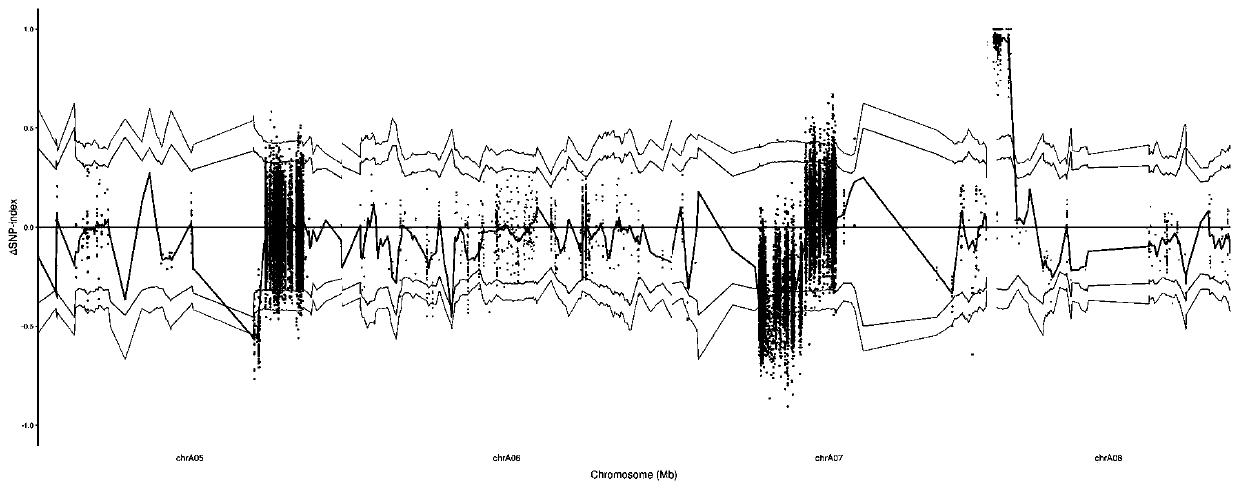
[0060] The parental materials are the homozygous dimorphic line D0721, the temporary protection line D1577 and the restorer line D0936 grown in the Zhuanghang Experimental Base of the Institute of Crop Breeding and Cultivation, Shanghai Academy of Agricultural Sciences.
[0061] 1) The sterile plant of the homozygous two-type line D0721 of Brassica napus was used as the mother, and the temporary insurance line D1577 was used as the male parent, and crossed to obtain a complete sterile line, a complete sterile line and a restorer line D0936 with no wax powder traits. Hybridization to obtain the F1 generation;
[0062] 3) From the 15:1 segregation population of the F2-F5 generation, the plants with the wax powder trait were selected for selfing to obtain the F6 generation.
Embodiment 2
[0064] 1 source of material
[0065] The test materials are the homozygous two-type line D0721, the temporary protection line D1577 and the restorer line D0936 planted in the Zhuanghang Experimental Base of the Institute of Crop Breeding and Cultivation of the Shanghai Academy of Agricultural Sciences. Wax plants and all with the wax phenotype did not segregate populations.
[0066] 2 methods
[0067] 2.1 Library construction and sequencing
[0068] The genomic DNA in the sample is extracted, and the library is constructed after the electrophoresis test is qualified. Qualified DNA samples were first randomly fragmented into 350bp fragments by Covaris, and the TruSeq DNA LT Sample Prepkit kit was used to build a library. The DNA fragments underwent end repair, poly(A) tailing, sequencing adapters, purification, and PCR amplification. and other steps, and finally complete the library construction, and use the sequencer to perform double-end sequencing after the library is qua...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com