Specific detection method of human breast cancer cells MCF-7 based on surface-enhanced Raman spectroscopy
A surface-enhanced Raman, human breast cancer cell technology, applied in Raman scattering, material excitation analysis, etc., can solve the problems of cell destruction and invasiveness, and achieve strong specificity, high sensitivity, and high Raman signal enhancement ability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
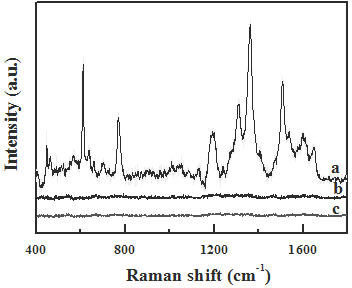
Image
Examples
Embodiment 1
[0032] Dissolve the aptamer (S2.2, sequence 5'-GCA GTT GAT CCT TTG GAT ACC CTG G-3') in 0.1 mol / L phosphate buffer (PBS, pH 7.4) at a concentration of 300 ng / mL ; the NaAuCl of 1mol / L 4 and 1mol / L AgNO 3 After the solution is mixed evenly at a volume ratio of 1:1, it is added to the nucleic acid aptamer solution, and the Au 3+ 、Ag + The mass ratio of base and base is 1:1:5. Stir the mixture continuously at 4°C for 0.5–2 hours, store it in the dark for 12 hours, and transfer it to a 1cm×1cm quartz cuvette In this method, the Au-Ag-aptamer nanocomposite was obtained by irradiating for 30 minutes under a UV lamp with a wave number of 254 nm; a 0.1 mol / L Rh 6G methanol solution was prepared and added to the Au-Ag-aptamer nanocomposite dispersion In the solution, control the final concentration of Rh 6G to 1 μmol / L, stir at 4 °C for 1 h in the dark, centrifuge at 10,000 rpm for 10 min, and wash the precipitate with 0.1 mol / L PBS solution (pH 7.4) for a total of 3 times , to rem...
Embodiment 2
[0034] Dissolve the aptamer (S2.2, sequence 5'-GCA GTT GAT CCT TTG GAT ACC CTG G-3') in 0.1 mol / L phosphate buffer (PBS, pH 7.4) at a concentration of 300 ng / mL ; the NaAuCl of 1mol / L 4 and 1mol / L AgNO 3 After the solution is mixed uniformly at a volume ratio of 2:1, it is added to the nucleic acid aptamer solution, and the Au 3+ 、Ag + The mass ratio of base and base is 2:1:5, and the mixture is continuously stirred at 4°C for 0.5–2 hours, and stored in a dark place away from light for 12 hours, and then transferred to a 1cm×1cm quartz cuvette In this method, the Au-Ag-aptamer nanocomposite was obtained by irradiating for 30 minutes under a UV lamp with a wave number of 254 nm; a 0.1 mol / L Rh 6G methanol solution was prepared and added to the Au-Ag-aptamer nanocomposite dispersion In the solution, control the final concentration of Rh 6G to 1 μmol / L, stir at 4 °C for 1 h in the dark, centrifuge at 10,000 rpm for 10 min, and wash the precipitate with 0.1 mol / L PBS solution (...
Embodiment 3
[0036] Dissolve the aptamer (S2.2, sequence 5'-GCA GTT GAT CCT TTG GAT ACC CTG G-3') in 0.1 mol / L phosphate buffer (PBS, pH 7.4) at a concentration of 300 ng / mL ; the NaAuCl of 1mol / L 4 and 1mol / L AgNO 3 After the solution is mixed uniformly at a volume ratio of 4:1, it is added to the nucleic acid aptamer solution, and the Au 3+ 、Ag + The mass ratio of base and base is 4:1:5. Stir the mixture continuously at 4°C for 0.5–2 hours, and store it in the dark for 12 hours, and transfer it to a 1cm×1cm quartz cuvette In this method, the Au-Ag-aptamer nanocomposite was obtained by irradiating for 30 minutes under a UV lamp with a wave number of 254 nm; a 0.1 mol / L Rh 6G methanol solution was prepared and added to the Au-Ag-aptamer nanocomposite dispersion In the solution, control the final concentration of Rh 6G to 1 μmol / L, stir at 4 °C for 1 h in the dark, centrifuge at 10,000 rpm for 10 min, and wash the precipitate with 0.1 mol / L PBS solution (pH 7.4) for a total of 3 times ,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com