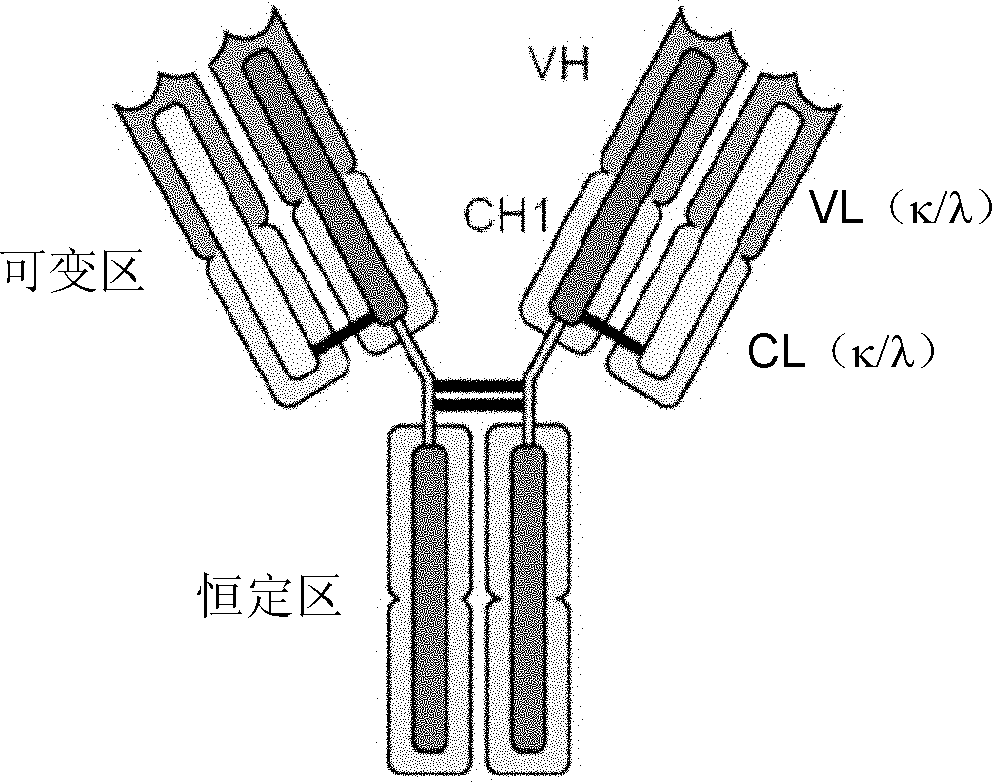
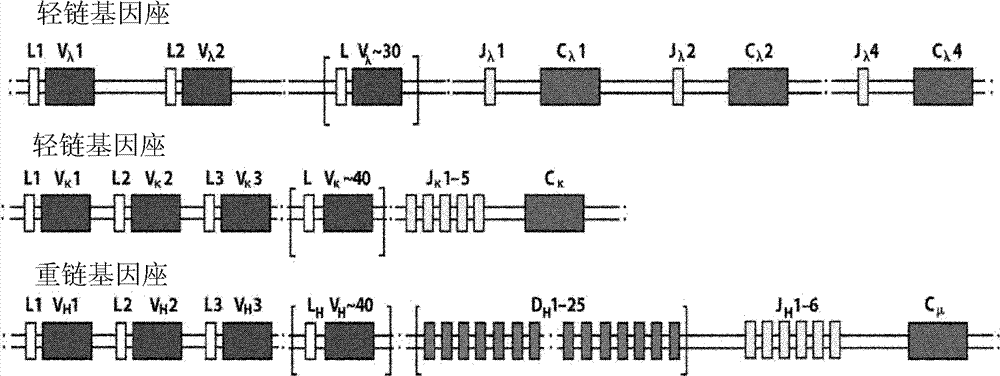
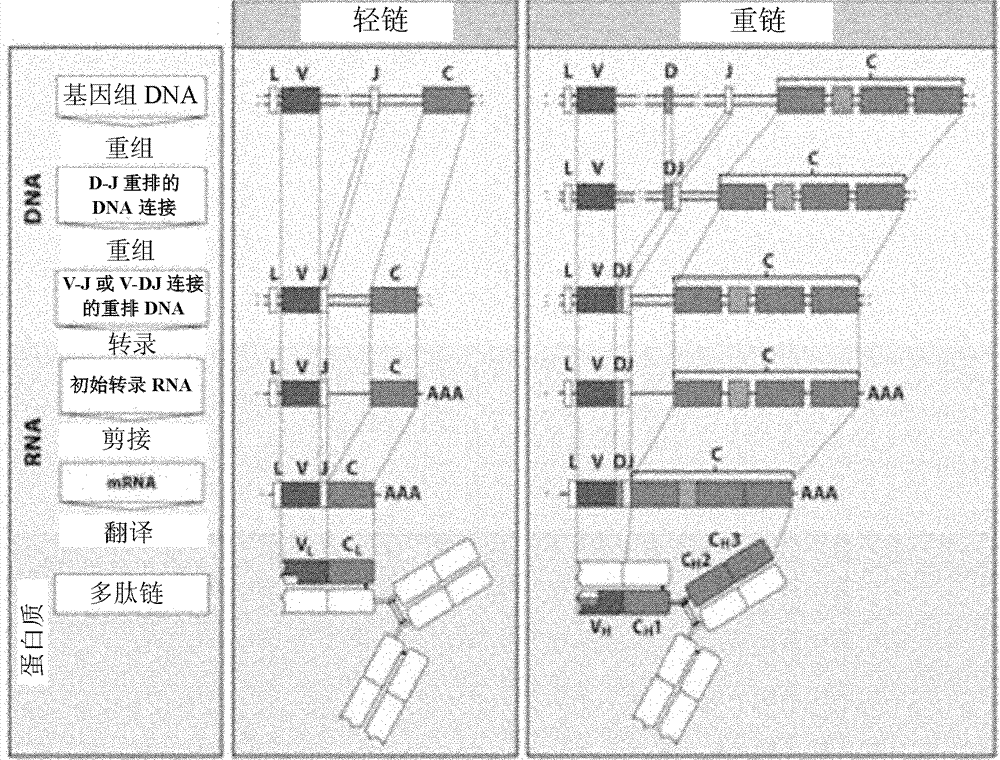
Method for rearranging immunoglobulin and producing antibody through MDA-PCR (Multiple Displacement Amplification-Polymerase Chain Reaction) enriched genome
An immunoglobulin and genome technology, applied in biochemical equipment and methods, genetic engineering, DNA preparation, etc., can solve problems such as substrates that are not suitable for PCR amplification of target genes, avoid mRNA instability, and reduce subsequent pollution. , the effect of simple steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Figure 5A The analyzes of lanes 1-5 and lanes 6-10 were for serial dilutions of mouse B cells (approximately 1000, 100, 10, 1, 0.1 cells). After cell lysis (step 1), VH (390-400bp) and Vk (370-380bp) genes were enriched with or without MDA (step 2), and then amplified by PCR (step 3), wherein lane 1- 5 performed PCR only, lanes 6-10 performed MDA-PCR as Figure 5A As shown, the VH and Vk genes can be simultaneously amplified in the reaction tube with only a single cell in each tube (channel 9), and if no MDA enrichment is carried out, the IgH gene can be detected only at the level of 100 and 1000 cells (channel 9). 2 and 1). in Figure 5A of channels 6-10 and Figure 5B The primers for MDA in lanes 6-10 are all the specific primers in Table 2. Figure 5A Lanes 1-5 are the results of direct PCR without MDA amplification, Figure 5B Lanes 1-5 are the results of PCR after MDA amplification with random hexamer primers. exist Figure 5B Lanes 6-10 show MDA enrichmen...
Embodiment 2
[0076] Example 2 is the isolation of mouse antibody genes from single B cells by MDA-PCR. Using fluorescence-activated cell sorting (FACS), single mouse κ memory B cells were distributed in 96-well plates and then enriched for MDA. respectively by IgH( Figure 6 , upper part) or Igk ( Figure 6 , lower) primers for further PCR amplification of MDA-enriched genomic DNA. Figure 6 Including upper and lower glue, the channel is corresponding. The upper gel shows the heavy chain (IgH) PCR amplification results of samples 1-24, and the lower gel shows the light chain (Igk) PCR amplification results of samples 1-24. The heavy and light chains of samples 1, 6, 10, 11, 12, 17, 20, and 21 (the samples indicated by the arrows) all obtained amplified target fragments, indicating that the samples were successfully amplified with complete antibodies Genes (both heavy and light chains), those samples where only one chain was amplified, were not actually able to obtain a complete antibod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com